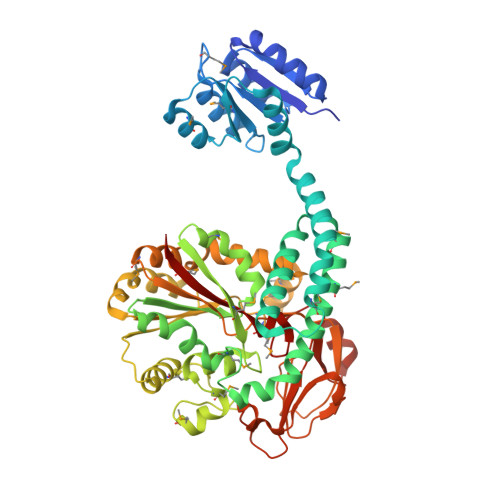
Response regulator PorX coordinates oligonucleotide signalling and gene expression to control the secretion of virulence factors.
Schmitz, C., Madej, M., Nowakowska, Z., Cuppari, A., Jacula, A., Ksiazek, M., Mikruta, K., Wisniewski, J., Pudelko-Malik, N., Saran, A., Zeytuni, N., Mlynarz, P., Lamont, R.J., Uson, I., Siksnys, V., Potempa, J., Sola, M.(2022) Nucleic Acids Res 50: 12558-12577
- PubMed: 36464236
- DOI: https://doi.org/10.1093/nar/gkac1103
- Primary Citation of Related Structures:
7PV7, 7PVA - PubMed Abstract:
The PglZ family of proteins belongs to the alkaline phosphatase superfamily, which consists of metallohydrolases with limited sequence identity but similar metal-coordination architectures in otherwise divergent active sites. Proteins with a well-defined PglZ domain are ubiquitous among prokaryotes as essential components of BREX phage defence systems and two-component systems (TCSs). Whereas other members of the alkaline phosphatase superfamily are well characterized, the activity, structure and biological function of PglZ family proteins remain unclear. We therefore investigated the structure and function of PorX, an orphan response regulator of the Porphyromonas gingivalis TCS containing a putative PglZ effector domain. The crystal structure of PorX revealed a canonical receiver domain, a helical bundle, and an unprecedented PglZ domain, similar to the general organization of the phylogenetically related BREX-PglZ proteins. The PglZ domain of PorX features an active site cleft suitable for large substrates. An extensive search for substrates revealed that PorX is a phosphodiesterase that acts on cyclic and linear oligonucleotides, including signalling molecules such as cyclic oligoadenylates. These results, combined with mutagenesis, biophysical and enzymatic analysis, suggest that PorX coordinates oligonucleotide signalling pathways and indirectly regulates gene expression to control the secretion of virulence factors.
- Department of Structural Biology, Molecular Biology Institute of Barcelona, CSIC, Barcelona Science Park, Barcelona E-08028, Spain.
Organizational Affiliation: