Crystal structure of the RNA-recognition motif of Drosophila melanogaster tRNA (uracil-5-)-methyltransferase homolog A
Witzenberger, M., Janowski, R., Niessing, D.(2024) Acta Crystallogr F Struct Biol Commun 80
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2024) Acta Crystallogr F Struct Biol Commun 80
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| FI05218p | 81 | Drosophila melanogaster | Mutation(s): 0 EC: 2.1.1.35 | 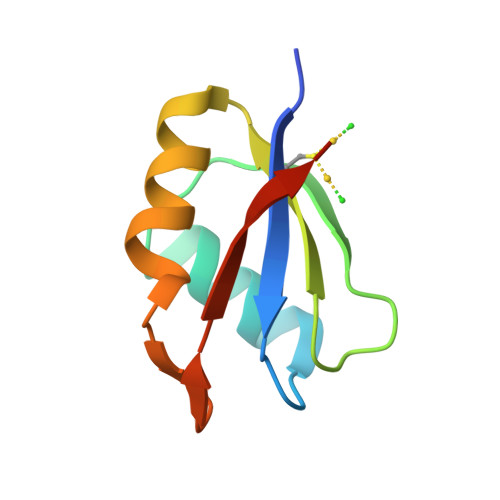 | |
UniProt | |||||
Find proteins for Q9VVV7 (Drosophila melanogaster) Explore Q9VVV7 Go to UniProtKB: Q9VVV7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9VVV7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | C [auth A], CA [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| CL Query on CL | D [auth A] DA [auth B] E [auth A] EA [auth B] F [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | AA [auth A] AB [auth B] BA [auth A] BB [auth B] CB [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| 85L Query on 85L | A, B | L-PEPTIDE LINKING | C3 H6 Au2 Cl2 N O2 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 92.41 | α = 90 |
| b = 41.89 | β = 110.8 |
| c = 48.82 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| XSCALE | data scaling |
| Auto-Rickshaw | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| German Research Foundation (DFG) | Germany | -- |