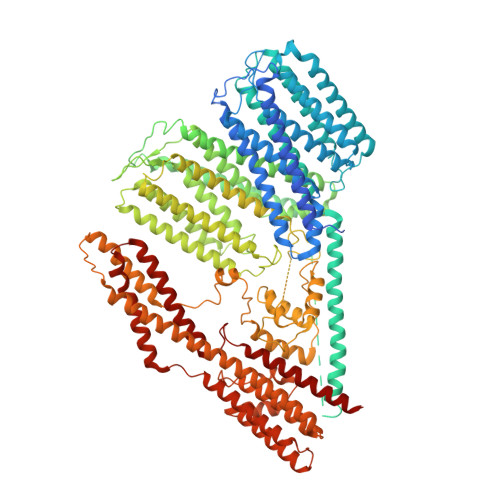
Rhodopsin-bestrophin fusion proteins from unicellular algae form gigantic pentameric ion channels.
Rozenberg, A., Kaczmarczyk, I., Matzov, D., Vierock, J., Nagata, T., Sugiura, M., Katayama, K., Kawasaki, Y., Konno, M., Nagasaka, Y., Aoyama, M., Das, I., Pahima, E., Church, J., Adam, S., Borin, V.A., Chazan, A., Augustin, S., Wietek, J., Dine, J., Peleg, Y., Kawanabe, A., Fujiwara, Y., Yizhar, O., Sheves, M., Schapiro, I., Furutani, Y., Kandori, H., Inoue, K., Hegemann, P., Beja, O., Shalev-Benami, M.(2022) Nat Struct Mol Biol 29: 592-603
- PubMed: 35710843
- DOI: https://doi.org/10.1038/s41594-022-00783-x
- Primary Citation of Related Structures:
7PL9 - PubMed Abstract:
Many organisms sense light using rhodopsins, photoreceptive proteins containing a retinal chromophore. Here we report the discovery, structure and biophysical characterization of bestrhodopsins, a microbial rhodopsin subfamily from marine unicellular algae, in which one rhodopsin domain of eight transmembrane helices or, more often, two such domains in tandem, are C-terminally fused to a bestrophin channel. Cryo-EM analysis of a rhodopsin-rhodopsin-bestrophin fusion revealed that it forms a pentameric megacomplex (~700 kDa) with five rhodopsin pseudodimers surrounding the channel in the center. Bestrhodopsins are metastable and undergo photoconversion between red- and green-absorbing or green- and UVA-absorbing forms in the different variants. The retinal chromophore, in a unique binding pocket, photoisomerizes from all-trans to 11-cis form. Heterologously expressed bestrhodopsin behaves as a light-modulated anion channel.
- Faculty of Biology, Technion - Israel Institute of Technology, Haifa, Israel.
Organizational Affiliation: