Mechanistic insights into the rational design of masked antibodies.
Orozco, C.T., Bersellini, M., Irving, L.M., Howard, W.W., Hargreaves, D., Devine, P.W.A., Siouve, E., Browne, G.J., Bond, N.J., Phillips, J.J., Ravn, P., Jackson, S.E.(null) MAbs 14: 2095701-2095701
- PubMed: 35799328
- DOI: https://doi.org/10.1080/19420862.2022.2095701
- Primary Citation of Related Structures:
7PKL - PubMed Abstract:
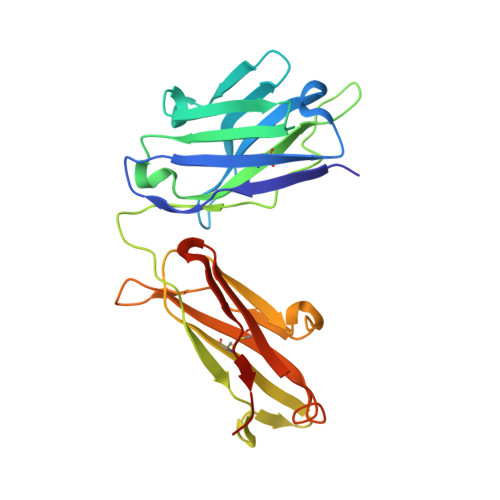
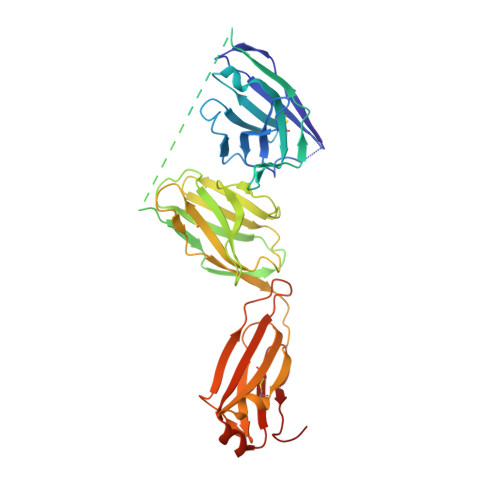
Although monoclonal antibodies have greatly improved cancer therapy, they can trigger side effects due to on-target, off-tumor toxicity. Over the past decade, strategies have emerged to successfully mask the antigen-binding site of antibodies, such that they are only activated at the relevant site, for example, after proteolytic cleavage. However, the methods for designing an ideal affinity-based mask and what parameters are important are not yet well understood. Here, we undertook mechanistic studies using three masks with different properties and identified four critical factors: binding site and affinity, as well as association and dissociation rate constants, which also played an important role. HDX-MS was used to identify the location of binding sites on the antibody, which were subsequently validated by obtaining a high-resolution crystal structure for one of the mask-antibody complexes. These findings will inform future designs of optimal affinity-based masks for antibodies and other therapeutic proteins.
- Yusuf Hamied Department of Chemistry, University of Cambridge, Cambridge, UK.
Organizational Affiliation: