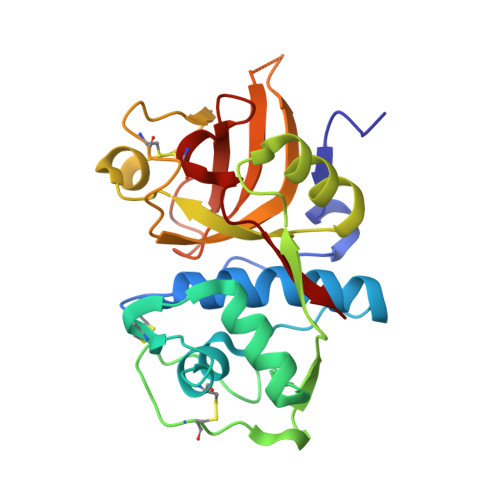
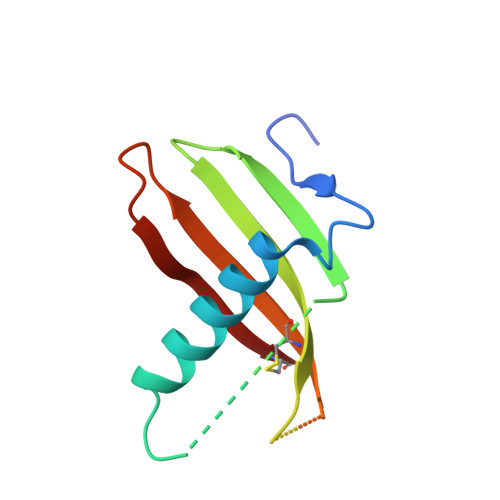
Protease-bound structure of Ricistatin provides insights into the mechanism of action of tick salivary cystatins in the vertebrate host.
Martins, L.A., Busa, M., Chlastakova, A., Kotal, J., Berankova, Z., Stergiou, N., Jmel, M.A., Schmitt, E., Chmelar, J., Mares, M., Kotsyfakis, M.(2023) Cell Mol Life Sci 80: 339-339
- PubMed: 37898573
- DOI: https://doi.org/10.1007/s00018-023-04993-4
- Primary Citation of Related Structures:
7PK4 - PubMed Abstract:
Tick saliva injected into the vertebrate host contains bioactive anti-proteolytic proteins from the cystatin family; however, the molecular basis of their unusual biochemical and physiological properties, distinct from those of host homologs, is unknown. Here, we present Ricistatin, a novel secreted cystatin identified in the salivary gland transcriptome of Ixodes ricinus ticks. Recombinant Ricistatin inhibited host-derived cysteine cathepsins and preferentially targeted endopeptidases, while having only limited impact on proteolysis driven by exopeptidases. Determination of the crystal structure of Ricistatin in complex with a cysteine cathepsin together with characterization of structural determinants in the Ricistatin binding site explained its restricted specificity. Furthermore, Ricistatin was potently immunosuppressive and anti-inflammatory, reducing levels of pro-inflammatory cytokines IL-6, IL-1β, and TNF-α and nitric oxide in macrophages; IL-2 and IL-9 levels in Th9 cells; and OVA antigen-induced CD4 + T cell proliferation and neutrophil migration. This work highlights the immunotherapeutic potential of Ricistatin and, for the first time, provides structural insights into the unique narrow selectivity of tick salivary cystatins determining their bioactivity.
- Institute of Parasitology, Branišovská 1160/31, 37005, Ceske Budejovice, Czech Republic.
Organizational Affiliation: