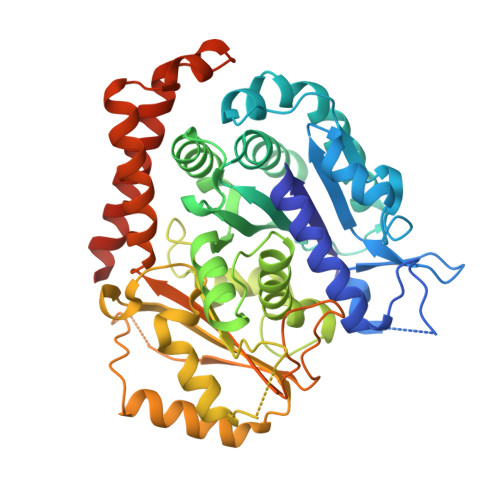
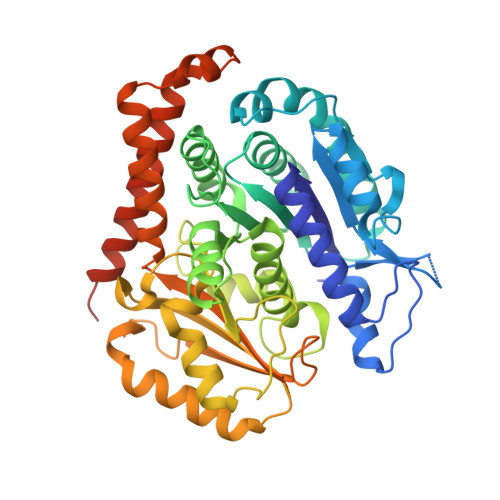
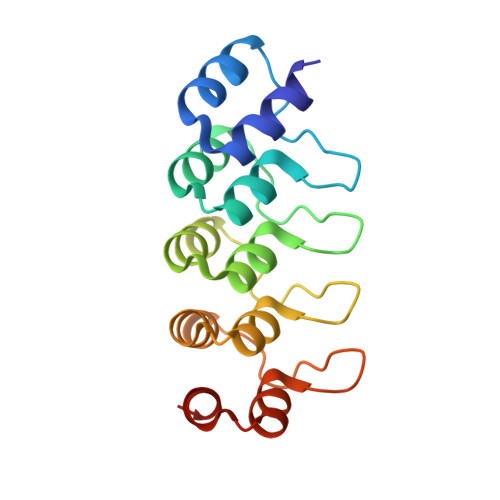
Inhibiting parasite proliferation using a rationally designed anti-tubulin agent.
Gaillard, N., Sharma, A., Abbaali, I., Liu, T., Shilliday, F., Cook, A.D., Ehrhard, V., Bangera, M., Roberts, A.J., Moores, C.A., Morrissette, N., Steinmetz, M.O.(2021) EMBO Mol Med 13: e13818-e13818
- PubMed: 34661376
- DOI: https://doi.org/10.15252/emmm.202013818
- Primary Citation of Related Structures:
7PJE, 7PJF - PubMed Abstract:
Infectious diseases caused by apicomplexan parasites remain a global public health threat. The presence of multiple ligand-binding sites in tubulin makes this protein an attractive target for anti-parasite drug discovery. However, despite remarkable successes as anti-cancer agents, the rational development of protozoan parasite-specific tubulin drugs has been hindered by a lack of structural and biochemical information on protozoan tubulins. Here, we present atomic structures for a protozoan tubulin and microtubule and delineate the architectures of apicomplexan tubulin drug-binding sites. Based on this information, we rationally designed the parasite-specific tubulin inhibitor parabulin and show that it inhibits growth of parasites while displaying no effects on human cells. Our work presents for the first time the rational design of a species-specific tubulin drug providing a framework to exploit structural differences between human and protozoa tubulin variants enabling the development of much-needed, novel parasite inhibitors.
- Laboratory of Biomolecular Research, Division of Biology and Chemistry, Paul Scherrer Institut, Villigen, Switzerland.
Organizational Affiliation: