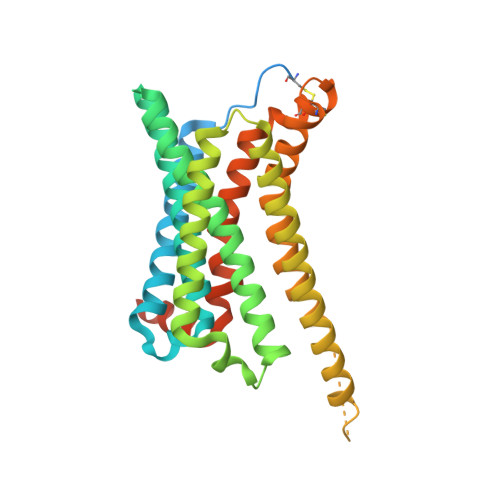
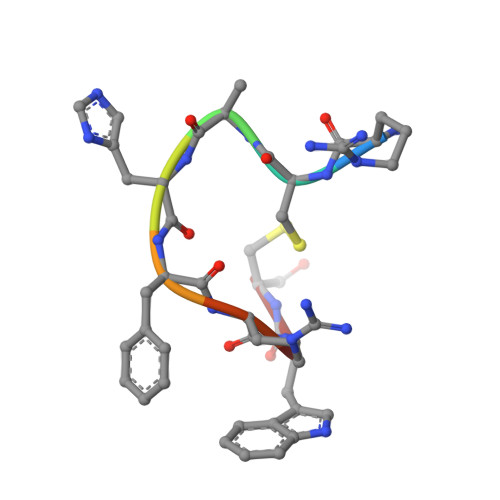
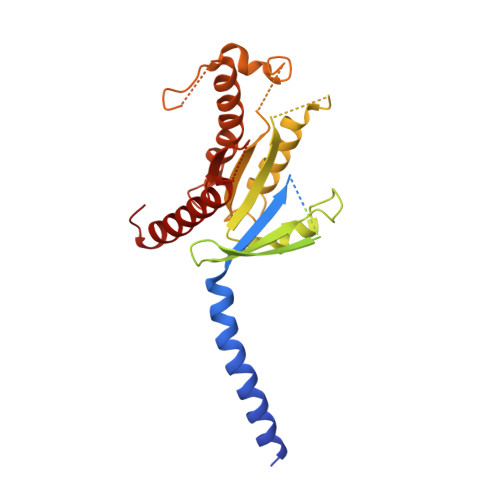
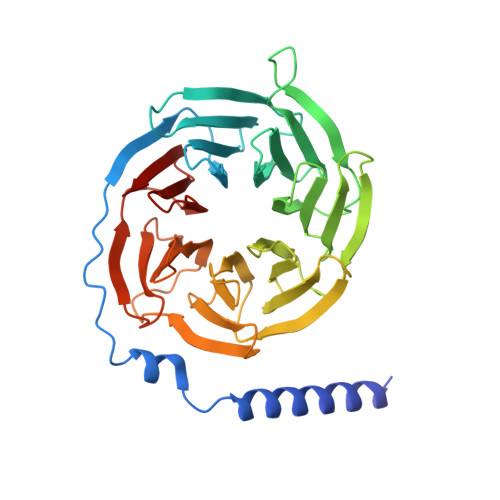
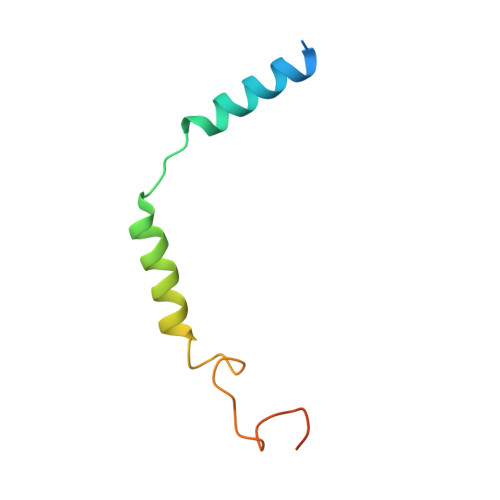
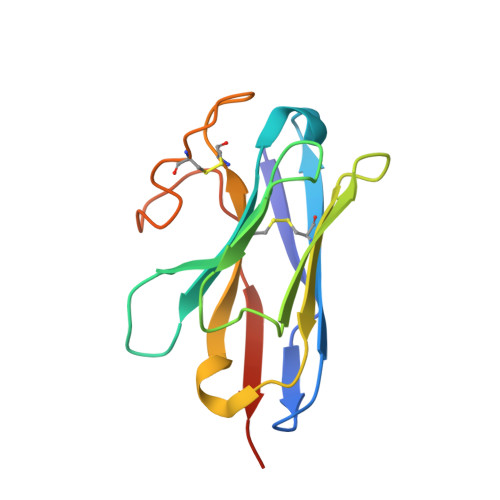
Structures of active melanocortin-4 receptor-Gs-protein complexes with NDP-alpha-MSH and setmelanotide.
Heyder, N.A., Kleinau, G., Speck, D., Schmidt, A., Paisdzior, S., Szczepek, M., Bauer, B., Koch, A., Gallandi, M., Kwiatkowski, D., Burger, J., Mielke, T., Beck-Sickinger, A.G., Hildebrand, P.W., Spahn, C.M.T., Hilger, D., Schacherl, M., Biebermann, H., Hilal, T., Kuhnen, P., Kobilka, B.K., Scheerer, P.(2021) Cell Res 31: 1176-1189
- PubMed: 34561620
- DOI: https://doi.org/10.1038/s41422-021-00569-8
- Primary Citation of Related Structures:
7PIU, 7PIV - PubMed Abstract:
The melanocortin-4 receptor (MC4R), a hypothalamic master regulator of energy homeostasis and appetite, is a class A G-protein-coupled receptor and a prime target for the pharmacological treatment of obesity. Here, we present cryo-electron microscopy structures of MC4R-Gs-protein complexes with two drugs recently approved by the FDA, the peptide agonists NDP-α-MSH and setmelanotide, with 2.9 Å and 2.6 Å resolution. Together with signaling data from structure-derived MC4R mutants, the complex structures reveal the agonist-induced origin of transmembrane helix (TM) 6-regulated receptor activation. The ligand-binding modes of NDP-α-MSH, a high-affinity linear variant of the endogenous agonist α-MSH, and setmelanotide, a cyclic anti-obesity drug with biased signaling toward Gq/11, underline the key role of TM3 in ligand-specific interactions and of calcium ion as a ligand-adaptable cofactor. The agonist-specific TM3 interplay subsequently impacts receptor-Gs-protein interfaces at intracellular loop 2, which also regulates the G-protein coupling profile of this promiscuous receptor. Finally, our structures reveal mechanistic details of MC4R activation/inhibition, and provide important insights into the regulation of the receptor signaling profile which will facilitate the development of tailored anti-obesity drugs.
- Charité - Universitätsmedizin Berlin, corporate member of Freie Universität Berlin and Humboldt-Universität zu Berlin, Institute of Medical Physics and Biophysics, Group Protein X-ray Crystallography and Signal Transduction, Charitéplatz 1, Berlin, Germany.
Organizational Affiliation: