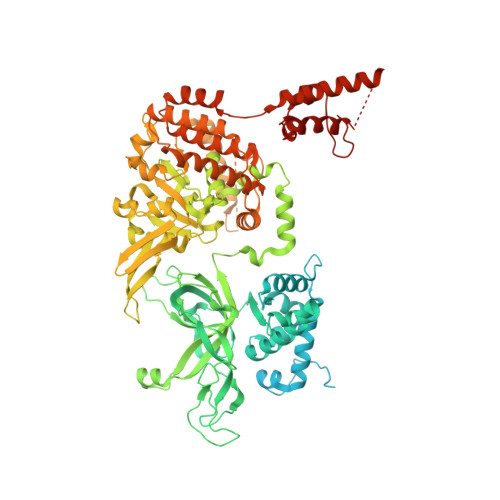
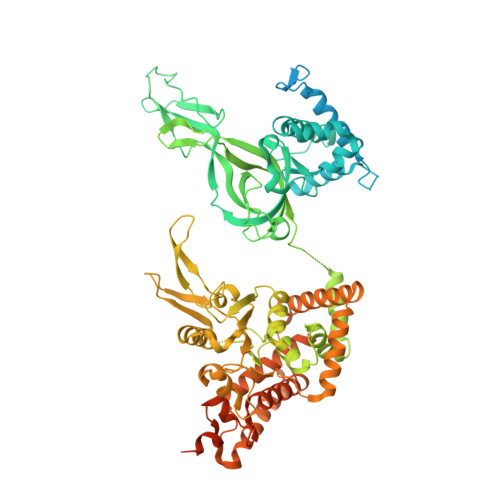
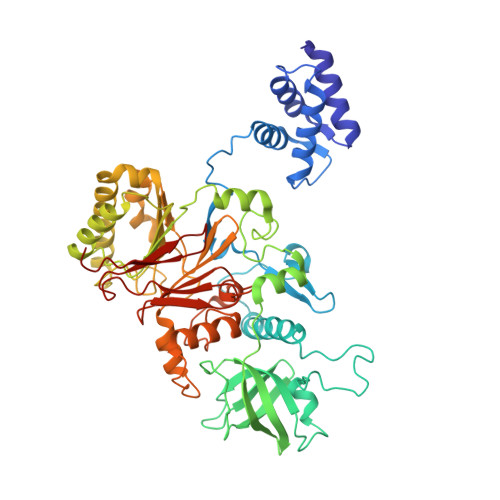
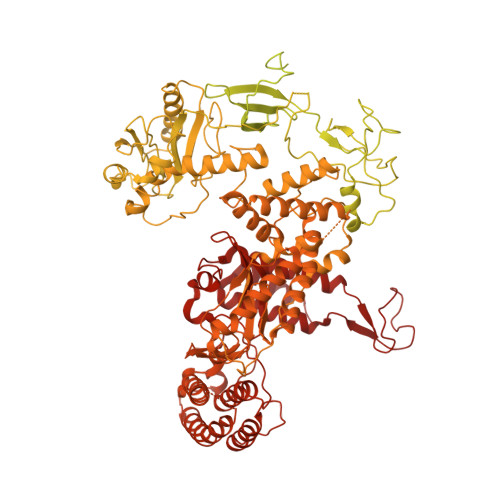
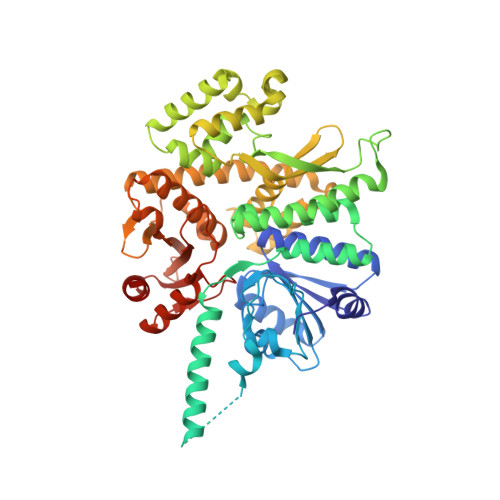
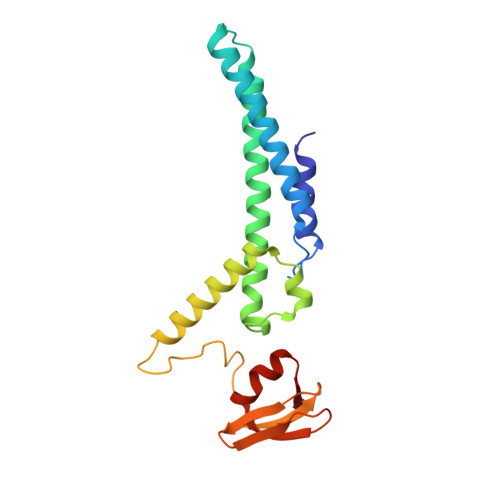
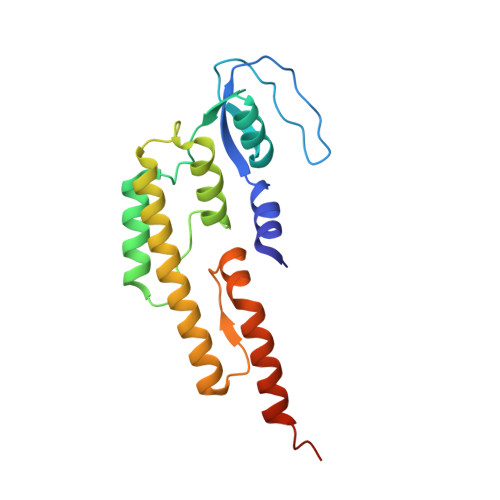
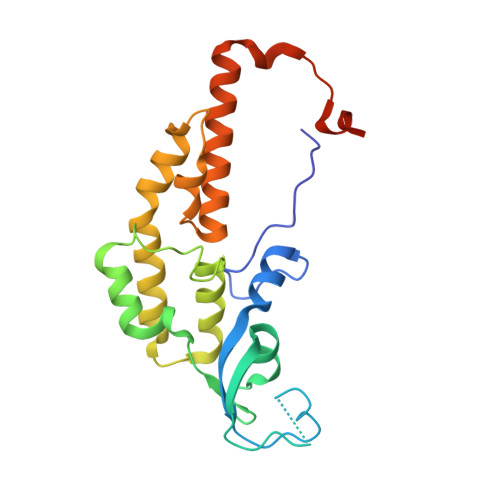
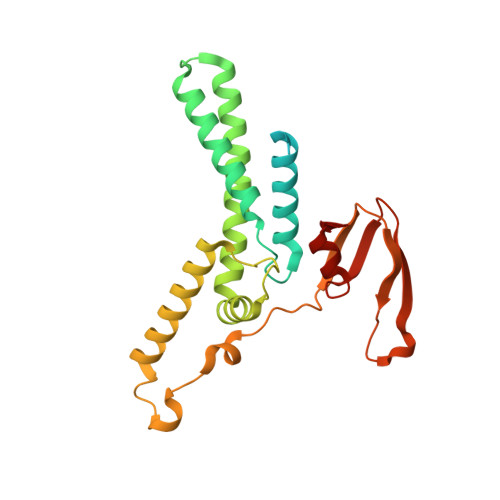
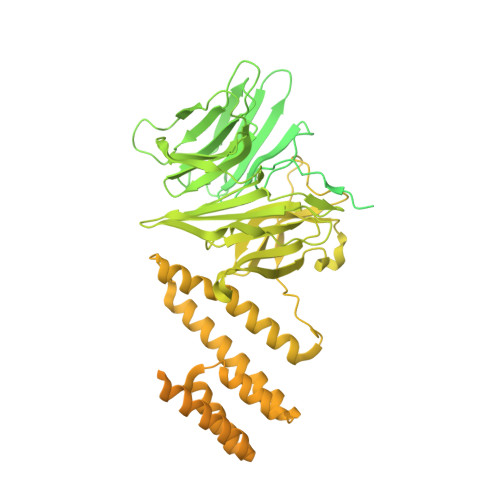
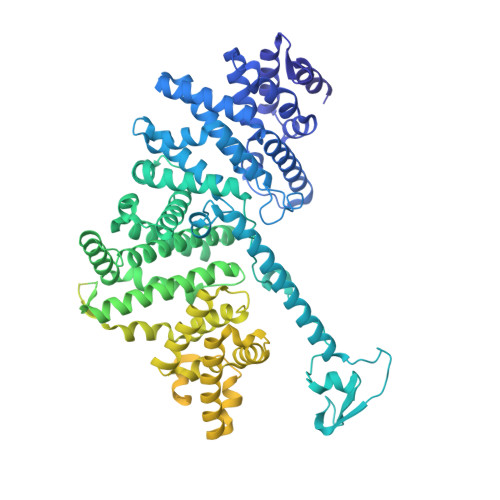
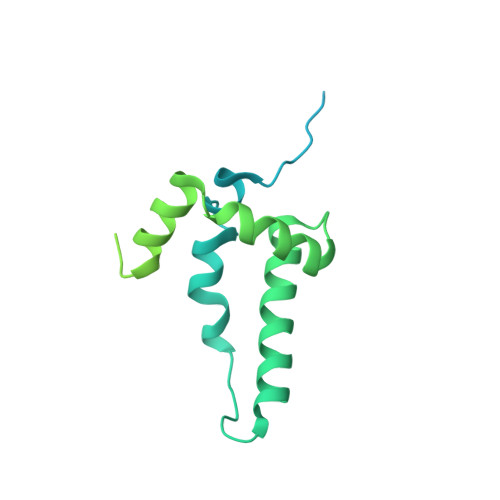
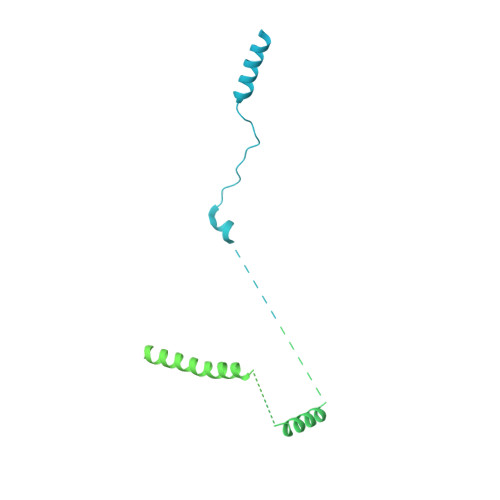
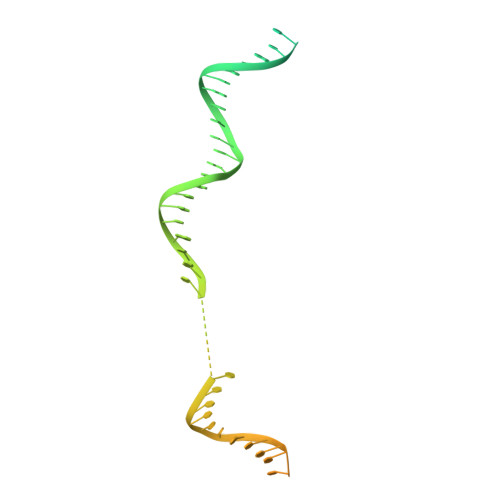
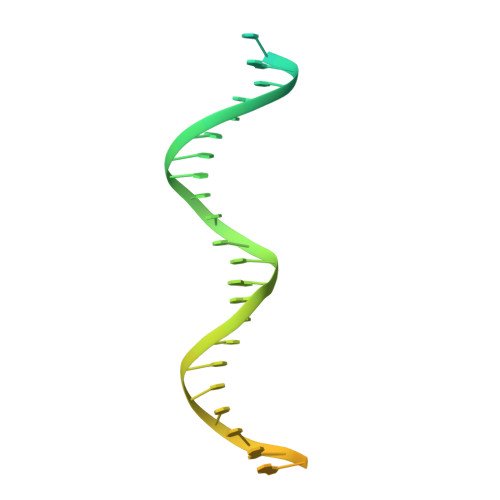
Structure of a human replisome shows the organisation and interactions of a DNA replication machine.
Jones, M.L., Baris, Y., Taylor, M.R.G., Yeeles, J.T.P.(2021) EMBO J 40: e108819-e108819
- PubMed: 34694004
- DOI: https://doi.org/10.15252/embj.2021108819
- Primary Citation of Related Structures:
7PFO - PubMed Abstract:
The human replisome is an elaborate arrangement of molecular machines responsible for accurate chromosome replication. At its heart is the CDC45-MCM-GINS (CMG) helicase, which, in addition to unwinding the parental DNA duplex, arranges many proteins including the leading-strand polymerase Pol ε, together with TIMELESS-TIPIN, CLASPIN and AND-1 that have key and varied roles in maintaining smooth replisome progression. How these proteins are coordinated in the human replisome is poorly understood. We have determined a 3.2 Å cryo-EM structure of a human replisome comprising CMG, Pol ε, TIMELESS-TIPIN, CLASPIN and AND-1 bound to replication fork DNA. The structure permits a detailed understanding of how AND-1, TIMELESS-TIPIN and Pol ε engage CMG, reveals how CLASPIN binds to multiple replisome components and identifies the position of the Pol ε catalytic domain. Furthermore, the intricate network of contacts contributed by MCM subunits and TIMELESS-TIPIN with replication fork DNA suggests a mechanism for strand separation.
- MRC Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: