Substrate Transport and Specificity in a Phospholipid Flippase
Wang, Y., Lyons, J.A., Timcenko, M., Kummerer, F., de Groot, B.L., Nissen, P., Gapsys, V., Lindorff-Larsen, K.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Probable phospholipid-transporting ATPase DRS2 | 1,355 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: DRS2, YAL026C, FUN38 EC: 7.6.2.1 Membrane Entity: Yes | 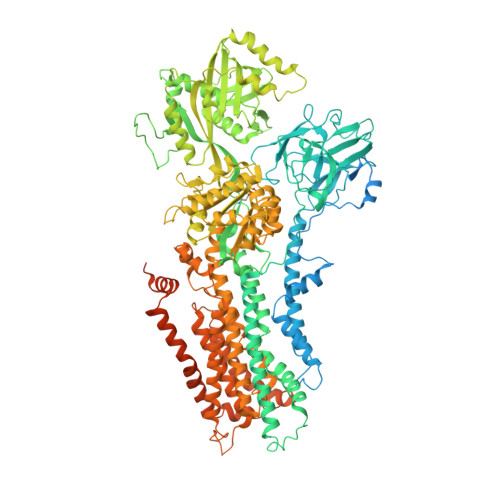 | |
UniProt | |||||
Find proteins for P39524 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P39524 Go to UniProtKB: P39524 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P39524 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cell division control protein 50 | B [auth C] | 391 | Saccharomyces cerevisiae S288C | Mutation(s): 0 Gene Names: CDC50, YCR094W, YCR94W Membrane Entity: Yes | 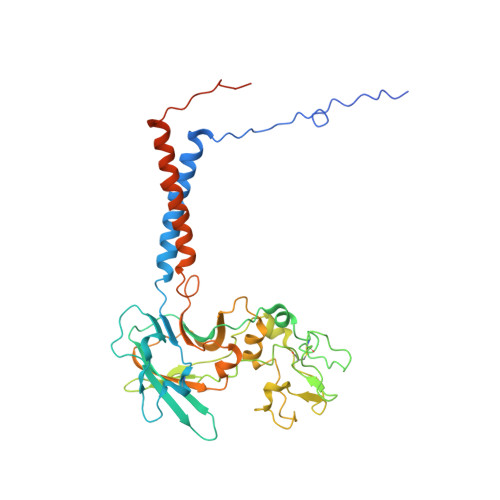 |
UniProt | |||||
Find proteins for P25656 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P25656 Go to UniProtKB: P25656 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P25656 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | Go to GlyGen: P25656-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | C [auth B], D | 2 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G42666HT GlyCosmos: G42666HT GlyGen: G42666HT | |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | E | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G31886NL GlyCosmos: G31886NL GlyGen: G31886NL | |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 2Y5 Query on 2Y5 | F [auth A] | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate C47 H84 O16 P2 ROKMWIUSHIHOGI-BWTMTLBLSA-N |  | ||
| Q3G (Subject of Investigation/LOI) Query on Q3G | I [auth A] | O-[(R)-[(2S)-2-(hexadecanoyloxy)-3-(octadecanoyloxy)propoxy](hydroxy)phosphoryl]-D-serine C40 H78 N O10 P WZFUPCSEUKNOBF-PQQNNWGCSA-N |  | ||
| ATP Query on ATP | H [auth A] | ADENOSINE-5'-TRIPHOSPHATE C10 H16 N5 O13 P3 ZKHQWZAMYRWXGA-KQYNXXCUSA-N |  | ||
| MG Query on MG | G [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| PHD Query on PHD | A | L-PEPTIDE LINKING | C4 H8 N O7 P |  | ASP |
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 3 |
| MODEL REFINEMENT | PHENIX |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Lundbeckfonden | Denmark | R155-2015-266 |