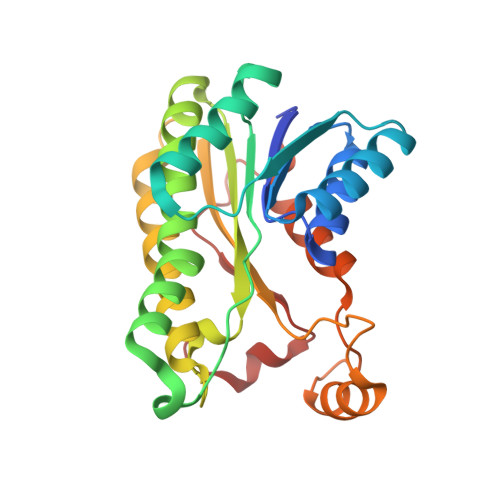
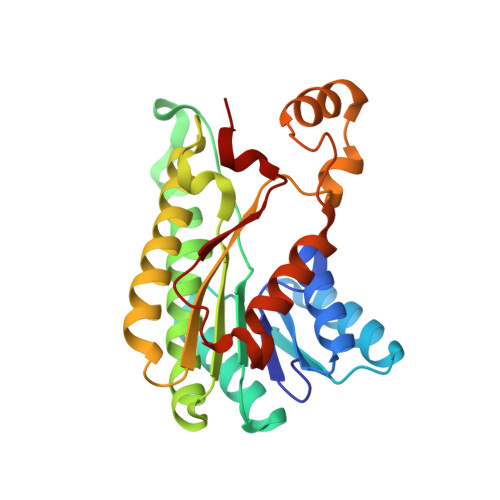
Inactive pseudoenzyme subunits in heterotetrameric BbsCD, a novel short-chain alcohol dehydrogenase involved in anaerobic toluene degradation.
von Horsten, S., Lippert, M.L., Geisselbrecht, Y., Schuhle, K., Schall, I., Essen, L.O., Heider, J.(2022) FEBS J 289: 1023-1042
- PubMed: 34601806
- DOI: https://doi.org/10.1111/febs.16216
- Primary Citation of Related Structures:
7PCS - PubMed Abstract:
Anaerobic toluene degradation proceeds by fumarate addition to produce (R)-benzylsuccinate as first intermediate, which is further degraded via β-oxidation by five enzymes encoded in the conserved bbs operon. This study characterizes two enzymes of this pathway, (E)-benzylidenesuccinyl-CoA hydratase (BbsH), and (S,R)-2-(α-hydroxybenzyl)succinyl-CoA dehydrogenase (BbsCD) from Thauera aromatica. BbsH, a member of the enoyl-CoA hydratase family, converts (E)-benzylidenesuccinyl-CoA to 2-(α-hydroxybenzyl)succinyl-CoA and was subsequently used in a coupled enzyme assay with BbsCD, which belongs to the short-chain dehydrogenases/reductase (SDR) family. The BbsCD crystal structure shows a C2-symmetric heterotetramer consisting of BbsC 2 and BbsD 2 dimers. BbsD subunits are catalytically active and capable of binding NAD + and substrate, whereas BbsC subunits represent built-in pseudoenzyme moieties lacking all motifs of the SDR family required for substrate binding or catalysis. Molecular modeling studies predict that the active site of BbsD is specific for conversion of the (S,R)-diastereomer of 2-(α-hydroxybenzyl)succinyl-CoA to (S)-2-benzoylsuccinyl-CoA by hydride transfer to the re-face of nicotinamide adenine dinucleotide (NAD) + . Furthermore, BbsC subunits are not engaged in substrate binding and merely serve as scaffold for the BbsD dimer. BbsCD represents a novel clade of related enzymes within the SDR family, which adopt a heterotetrameric architecture and catalyze the β-oxidation of aromatic succinate adducts.
- Department of Chemistry, Philipps-Universität, Marburg, Germany.
Organizational Affiliation: