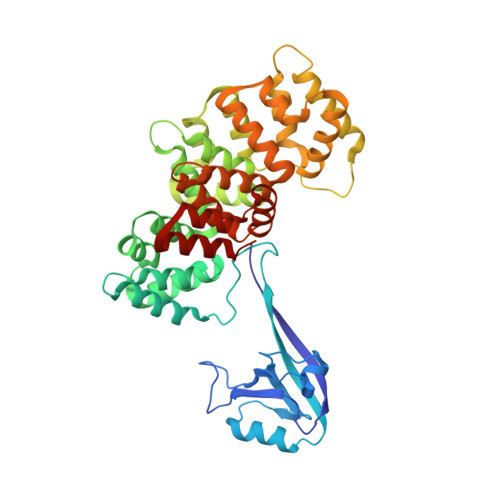
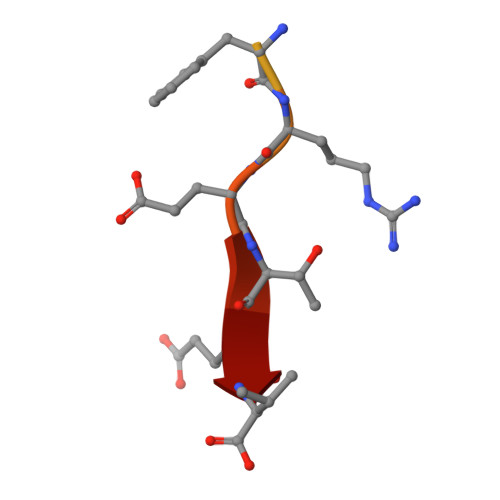
Quantitative fragmentomics allow affinity mapping of interactomes.
Gogl, G., Zambo, B., Kostmann, C., Cousido-Siah, A., Morlet, B., Durbesson, F., Negroni, L., Eberling, P., Jane, P., Nomine, Y., Zeke, A., Ostergaard, S., Monsellier, E., Vincentelli, R., Trave, G.(2022) Nat Commun 13: 5472-5472
- PubMed: 36115835
- DOI: https://doi.org/10.1038/s41467-022-33018-0
- Primary Citation of Related Structures:
7P70, 7P71, 7P72, 7P73, 7P74 - PubMed Abstract:
Human protein networks have been widely explored but most binding affinities remain unknown, hindering quantitative interactome-function studies. Yet interactomes rely on minimal interacting fragments displaying quantifiable affinities. Here, we measure the affinities of 65,000 interactions involving PDZ domains and their target PDZ-binding motifs (PBM) within a human interactome region particularly relevant for viral infection and cancer. We calculate interactomic distances, identify hot spots for viral interference, generate binding profiles and specificity logos, and explain selected cases by crystallographic studies. Mass spectrometry experiments on cell extracts and literature surveys show that quantitative fragmentomics effectively complements protein interactomics by providing affinities and completeness of coverage, putting a full human interactome affinity survey within reach. Finally, we show that interactome hijacking by the viral PBM of human papillomavirus E6 oncoprotein substantially impacts the host cell proteome beyond immediate E6 binders, illustrating the complex system-wide relationship between interactome and function.
- Équipe Labellisée Ligue 2015, Département de Biologie Structurale Intégrative, Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC), INSERM U1258/CNRS UMR 7104/Université de Strasbourg, 1 rue Laurent Fries, BP 10142, F-67404, Illkirch, France. goglg@igbmc.fr.
Organizational Affiliation: