Structure of the fungal hydroxylase, CYP505A30, and rational transfer of mutation data from CYP102A1 to alter regioselectivity
Aschenbrenner, J.C., Ebrecht, A.C., Tolmie, C., Smit, M.S., Opperman, D.J.(2021) Catal Sci Technol 11: 7359-7367
Experimental Data Snapshot
Starting Model: experimental
View more details
(2021) Catal Sci Technol 11: 7359-7367
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Bifunctional cytochrome P450/NADPH--P450 reductase | A [auth B], B [auth A], C, D | 464 | Thermothelomyces thermophilus ATCC 42464 | Mutation(s): 0 Gene Names: MYCTH_101224 EC: 1.14.14.1 (PDB Primary Data), 1.6.2.4 (PDB Primary Data) | 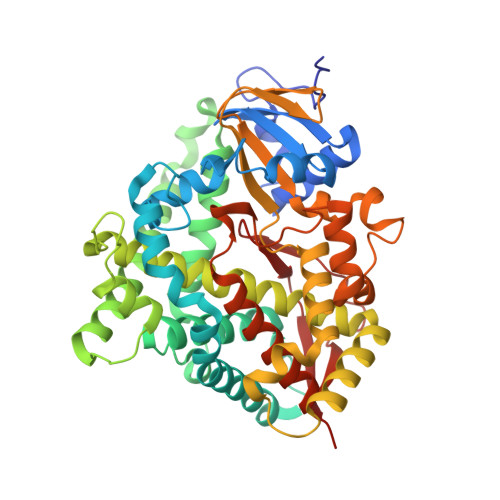 |
UniProt | |||||
Find proteins for G2QDZ3 (Thermothelomyces thermophilus (strain ATCC 42464 / BCRC 31852 / DSM 1799)) Explore G2QDZ3 Go to UniProtKB: G2QDZ3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | G2QDZ3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEM Query on HEM | E [auth B], G [auth A], I [auth C], K [auth D] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| DAO Query on DAO | F [auth B], H [auth A], J [auth C], L [auth D] | LAURIC ACID C12 H24 O2 POULHZVOKOAJMA-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 171.49 | α = 90 |
| b = 171.49 | β = 90 |
| c = 175.647 | γ = 90 |
| Software Name | Purpose |
|---|---|
| xia2 | data scaling |
| PHASER | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| DIALS | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Research Foundation in South Africa | South Africa | c*change, PAR-02 |
| National Research Foundation in South Africa | South Africa | 112094 |
| Global Challenges Research Fund | United Kingdom | ST/R002754/1 |