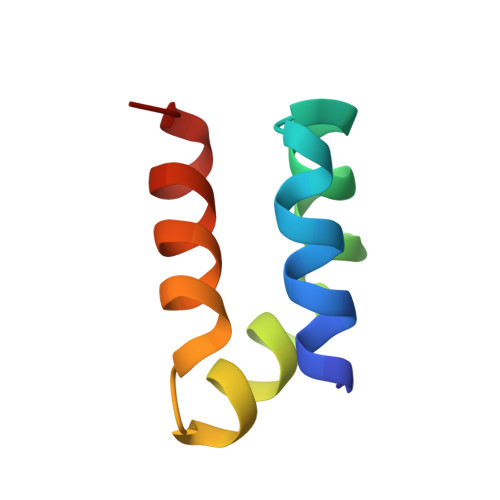
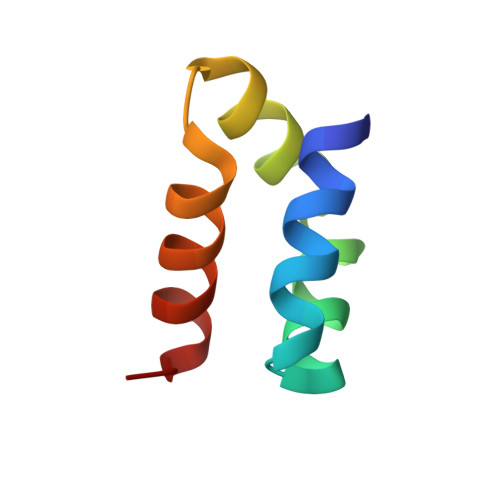Roles of inter- and intramolecular tryptophan interactions in membrane-active proteins revealed by racemic protein crystallography.
Lander, A.J., Mercado, L.D., Li, X., Taily, I.M., Findlay, B.L., Jin, Y., Luk, L.Y.P.(2023) Commun Chem 6: 154-154
- PubMed: 37464011
- DOI: https://doi.org/10.1038/s42004-023-00953-y
- Primary Citation of Related Structures:
7P5R, 8AVR, 8AVS, 8AVT, 8AVU - PubMed Abstract:
Tryptophan is frequently found on the surface of membrane-associated proteins that interact with the lipid membrane. However, because of their multifaceted interactions, it is difficult to pinpoint the structure-activity relationship of each tryptophan residue. Here, we describe the use of racemic protein crystallography to probe dedicated tryptophan interactions of a model tryptophan-rich bacteriocin aureocin A53 (AucA) by inclusion and/or exclusion of potential ligands. In the presence of tetrahedral anions that are isosteric to the head group of phospholipids, distinct tryptophan H-bond networks were revealed. H-bond donation by W40 was critical for antibacterial activity, as its substitution by 1-methyltryptophan resulted in substantial loss of activity against bacterial clinical isolates. Meanwhile, exclusion of tetrahedral ions revealed that W3 partakes in formation of a dimeric interface, thus suggesting that AucA is dimeric in solution and dissociated to interact with the phosphate head group in the presence of the lipid membrane. Based on these findings, we could predict the tryptophan residue responsible for activity as well as the oligomeric state of a distant homologue lacticin Q (48%).
- School of Chemistry, Cardiff University, Main Building, Park Place, Cardiff, CF10 3AT, UK.
Organizational Affiliation: