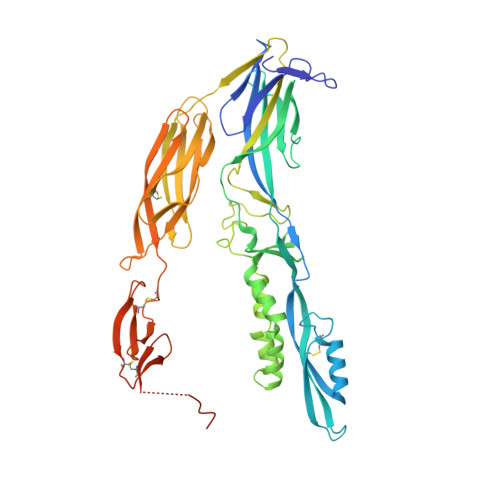
Discovery of archaeal fusexins homologous to eukaryotic HAP2/GCS1 gamete fusion proteins.
Moi, D., Nishio, S., Li, X., Valansi, C., Langleib, M., Brukman, N.G., Flyak, K., Dessimoz, C., de Sanctis, D., Tunyasuvunakool, K., Jumper, J., Grana, M., Romero, H., Aguilar, P.S., Jovine, L., Podbilewicz, B.(2022) Nat Commun 13: 3880-3880
- PubMed: 35794124
- DOI: https://doi.org/10.1038/s41467-022-31564-1
- Primary Citation of Related Structures:
7P4L - PubMed Abstract:
Sexual reproduction consists of genome reduction by meiosis and subsequent gamete fusion. The presence of genes homologous to eukaryotic meiotic genes in archaea and bacteria suggests that DNA repair mechanisms evolved towards meiotic recombination. However, fusogenic proteins resembling those found in gamete fusion in eukaryotes have so far not been found in prokaryotes. Here, we identify archaeal proteins that are homologs of fusexins, a superfamily of fusogens that mediate eukaryotic gamete and somatic cell fusion, as well as virus entry. The crystal structure of a trimeric archaeal fusexin (Fusexin1 or Fsx1) reveals an archetypical fusexin architecture with unique features such as a six-helix bundle and an additional globular domain. Ectopically expressed Fusexin1 can fuse mammalian cells, and this process involves the additional globular domain and a conserved fusion loop. Furthermore, archaeal fusexin genes are found within integrated mobile elements, suggesting potential roles in cell-cell fusion and gene exchange in archaea, as well as different scenarios for the evolutionary history of fusexins.
- Instituto de Fisiología, Biología Molecular y Neurociencias (IFIBYNE-CONICET), Buenos Aires, Argentina.
Organizational Affiliation: