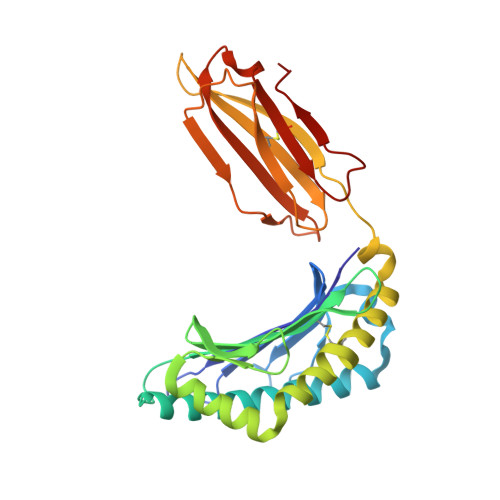
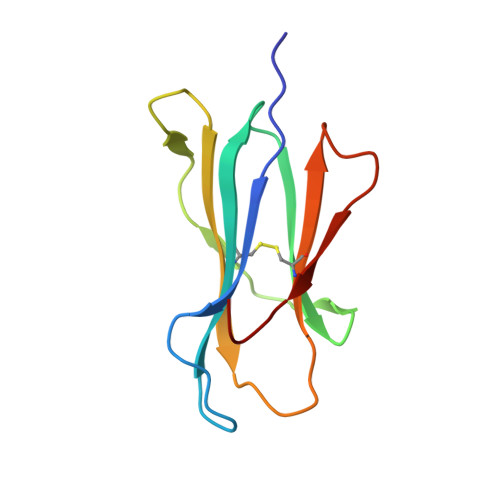
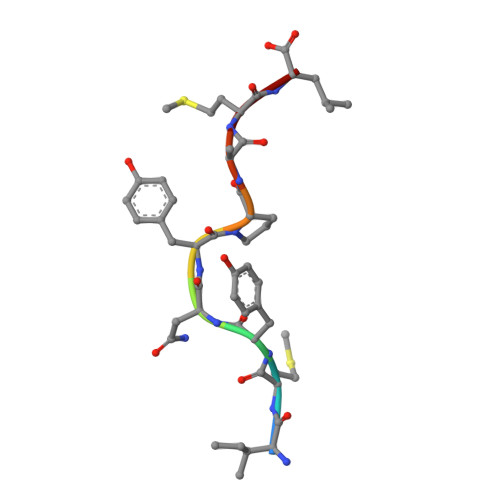
Primary and secondary functions of HLA-E are determined by stability and conformation of the peptide-bound complexes.
Walters, L.C., Rozbesky, D., Harlos, K., Quastel, M., Sun, H., Springer, S., Rambo, R.P., Mohammed, F., Jones, E.Y., McMichael, A.J., Gillespie, G.M.(2022) Cell Rep 39: 110959-110959
- PubMed: 35705051
- DOI: https://doi.org/10.1016/j.celrep.2022.110959
- Primary Citation of Related Structures:
7P49, 7P4B - PubMed Abstract:
MHC-E regulates NK cells by displaying MHC class Ia signal peptides (VL9) to NKG2A:CD94 receptors. MHC-E can also present sequence-diverse, lower-affinity, pathogen-derived peptides to T cell receptors (TCRs) on CD8 + T cells. To understand these affinity differences, human MHC-E (HLA-E)-VL9 versus pathogen-derived peptide structures are compared. Small-angle X-ray scatter (SAXS) measures biophysical parameters in solution, allowing comparison with crystal structures. For HLA-E-VL9, there is concordance between SAXS and crystal parameters. In contrast, HLA-E-bound pathogen-derived peptides produce larger SAXS dimensions that reduce to their crystallographic dimensions only when excess peptide is supplied. Further crystallographic analysis demonstrates three amino acids, exclusive to MHC-E, that not only position VL9 close to the α2 helix, but also allow non-VL9 peptide binding with re-configuration of a key TCR-interacting α2 region. Thus, non-VL9-bound peptides introduce an alternative peptide-binding motif and surface recognition landscape, providing a likely basis for VL9- and non-VL9-HLA-E immune discrimination.
- Nuffield Department of Medicine Research Building, Nuffield Department of Medicine, University of Oxford, Roosevelt Drive, Oxford OX3 7FZ, UK.
Organizational Affiliation: