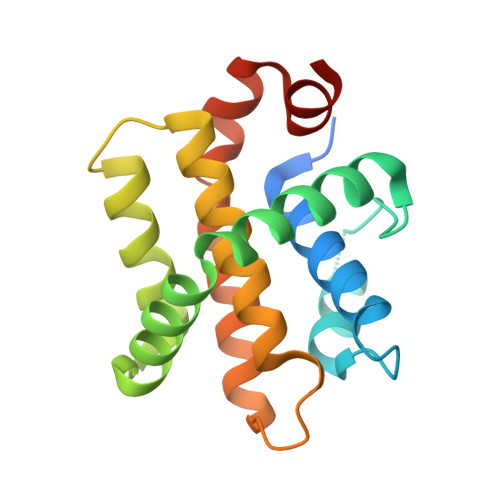
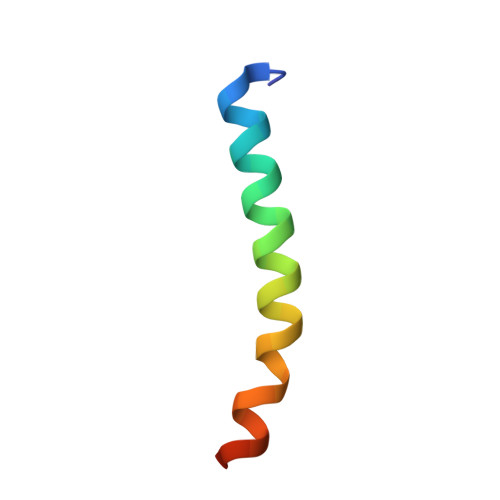
Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Suraweera, C.D., Hinds, M.G., Kvansakul, M.(2022) Viruses 14
- PubMed: 36298777
- DOI: https://doi.org/10.3390/v14102222
- Primary Citation of Related Structures:
7P33, 7P9W - PubMed Abstract:
Apoptosis is a powerful defense mechanism used by multicellular organisms to counteract viral infection. In response to premature host cell suicide, viruses have evolved numerous countermeasures to ensure cell viability to optimize their replication by encoding proteins homologous in structure and function to cellular pro-survival Bcl-2 proteins. Epstein-Barr virus (EBV), a member of the Gammaherpesviridae , encodes the Bcl-2 homolog BHRF1, a potent inhibitor of Bcl-2-mediated apoptosis. BHRF1 acts by directly targeting Bid and Puma, two proapoptotic proteins of the Bcl-2 family. Here, we determined the crystal structures of BHRF1 bound to peptides spanning the Bcl-2 binding motifs (Bcl-2 homology 3 motif, BH3) of Bid and Puma. BHRF1 engages BH3 peptides using the canonical ligand-binding groove of its Bcl-2 fold and maintains a salt bridge between an Arg residue with a conserved Asp residue in the BH3 motif mimicking the canonical ionic interaction seen in host Bcl-2:BH3 motif complexes. Furthermore, both Bid and Puma utilize a fifth binding pocket in the canonical ligand binding groove of BHRF1 to provide an additional hydrophobic interaction distinct from the interactions previously seen with Bak and Bim. These findings provide a structural basis for EBV-mediated suppression of host cell apoptosis and reveal the flexibility of virus encoded Bcl-2 proteins in mimicking key interactions from the endogenous host signaling pathways.
- Department of Biochemistry & Chemistry, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, VIC 3086, Australia.
Organizational Affiliation: