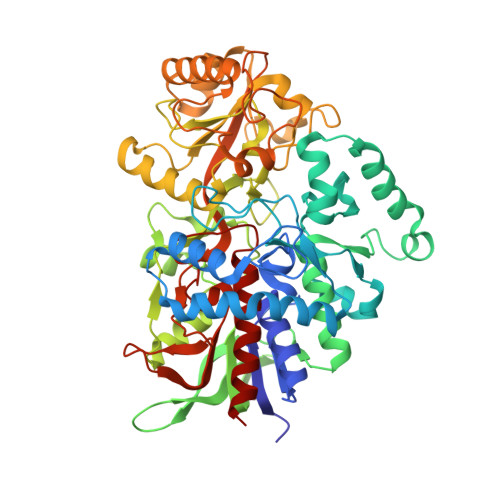
Structure, Mutagenesis, and QM:MM Modeling of 3-Ketosteroid Delta 1 -Dehydrogenase from Sterolibacterium denitrificans ─The Role of a New Putative Membrane-Associated Domain and Proton-Relay System in Catalysis.
Wojcik, P., Glanowski, M., Mrugala, B., Procner, M., Zastawny, O., Flejszar, M., Kurpiewska, K., Niedzialkowska, E., Minor, W., Oszajca, M., Bojarski, A.J., Wojtkiewicz, A.M., Szaleniec, M.(2023) Biochemistry 62: 808-823
- PubMed: 36625854
- DOI: https://doi.org/10.1021/acs.biochem.2c00576
- Primary Citation of Related Structures:
7P18 - PubMed Abstract:
3-Ketosteroid Δ 1 -dehydrogenases (KstD) are important microbial flavin enzymes that initiate the metabolism of steroid ring A and find application in the synthesis of steroid drugs. We present a structure of the KstD from Sterolibacterium denitrificans (AcmB), which contains a previously uncharacterized putative membrane-associated domain and extended proton-relay system. The experimental and theoretical studies show that the steroid Δ 1 -dehydrogenation proceeds according to the Ping-Pong bi-bi kinetics and a two-step base-assisted elimination (E2cB) mechanism. The mechanism is validated by evaluating the experimental and theoretical kinetic isotope effect for deuterium-substituted substrates. The role of the active-site residues is quantitatively assessed by point mutations, experimental activity assays, and QM/MM MD modeling of the reductive half-reaction (RHR). The pre-steady-state kinetics also reveals that the low pH (6.5) optimum of AcmB is dictated by the oxidative half-reaction (OHR), while the RHR exhibits a slight optimum at the pH usual for the KstD family of 8.5. The modeling confirms the origin of the enantioselectivity of C2-H activation and substrate specificity for Δ 4 -3-ketosteroids. Finally, the cholest-4-en-3-one turns out to be the best substrate of AcmB in terms of Δ G of binding and predicted rate of dehydrogenation.
- Jerzy Haber Institute of Catalysis and Surface Chemistry, Polish Academy of Sciences, Niezapominajek 8, 30-239Kraków, Poland.
Organizational Affiliation: