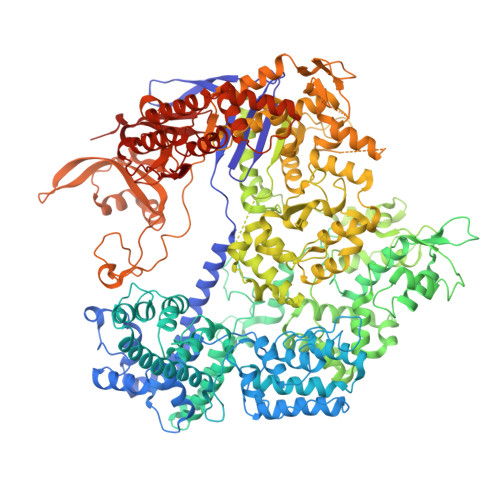
Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Donohoue, P.D., Pacesa, M., Lau, E., Vidal, B., Irby, M.J., Nyer, D.B., Rotstein, T., Banh, L., Toh, M.S., Gibson, J., Kohrs, B., Baek, K., Owen, A.L.G., Slorach, E.M., van Overbeek, M., Fuller, C.K., May, A.P., Jinek, M., Cameron, P.(2021) Mol Cell 81: 3637-3649.e5
- PubMed: 34478654
- DOI: https://doi.org/10.1016/j.molcel.2021.07.035
- Primary Citation of Related Structures:
7OX7, 7OX8, 7OX9, 7OXA - PubMed Abstract:
The off-target activity of the CRISPR-associated nuclease Cas9 is a potential concern for therapeutic genome editing applications. Although high-fidelity Cas9 variants have been engineered, they exhibit varying efficiencies and have residual off-target effects, limiting their applicability. Here, we show that CRISPR hybrid RNA-DNA (chRDNA) guides provide an effective approach to increase Cas9 specificity while preserving on-target editing activity. Across multiple genomic targets in primary human T cells, we show that 2'-deoxynucleotide (dnt) positioning affects guide activity and specificity in a target-dependent manner and that this can be used to engineer chRDNA guides with substantially reduced off-target effects. Crystal structures of DNA-bound Cas9-chRDNA complexes reveal distorted guide-target duplex geometry and allosteric modulation of Cas9 conformation. These structural effects increase specificity by perturbing DNA hybridization and modulating Cas9 activation kinetics to disfavor binding and cleavage of off-target substrates. Overall, these results pave the way for utilizing customized chRDNAs in clinical applications.
- Caribou Biosciences, Inc., 2929 Seventh Street, Suite 105, Berkeley, CA 94710, USA. Electronic address: donohoue@cariboubio.com.
Organizational Affiliation: