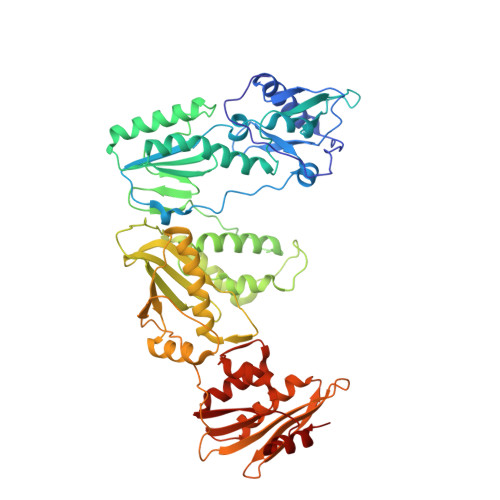
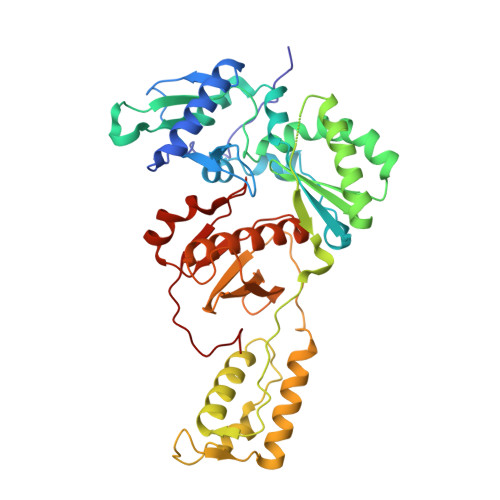
Exploring the dNTP -binding site of HIV-1 reverse transcriptase for inhibitor design.
Gu, W., Martinez, S., Singh, A.K., Nguyen, H., Rozenski, J., Schols, D., Herdewijn, P., Das, K., De Jonghe, S.(2021) Eur J Med Chem 225: 113785-113785
- PubMed: 34425311
- DOI: https://doi.org/10.1016/j.ejmech.2021.113785
- Primary Citation of Related Structures:
7OT6, 7OTA, 7OTK, 7OTN, 7OTX, 7OTZ, 7OUT - PubMed Abstract:
HIV-1 reverse transcriptase (RT) plays a central role in the viral life cycle, and roughly half of the FDA-approved anti-HIV drugs are targeting RT. Nucleoside analogs (NRTIs) require cellular phosphorylation for binding to RT, and to bypass this rate-limiting path, we designed a new series of acyclic nucleoside phosphonate analogs as nucleoside triphosphate mimics, aiming at the chelation of the catalytic Mg 2+ ions via a phosphonate and/or a carboxylic acid group. Novel synthetic procedures were developed to access these nucleoside phosphonate analogs. X-ray structures in complex with HIV-1 RT/dsDNA demonstrated that their binding modes are distinct from that of our previously reported compound series. The impact of chain length, chirality and linker atom have been discussed. The detailed structural understanding of these new compounds provides opportunities for designing new class of HIV-1 RT inhibitors.
- KU Leuven, Department of Microbiology, Immunology and Transplantation, Rega Institute for Medical Research, Laboratory of Virology and Chemotherapy, Herestraat 49, 3000, Leuven, Belgium; KU Leuven, Department of Pharmaceutical and Pharmacological Sciences, Rega Institute for Medical Research, Laboratory of Medicinal Chemistry, Herestraat 49, 3000, Leuven, Belgium.
Organizational Affiliation: