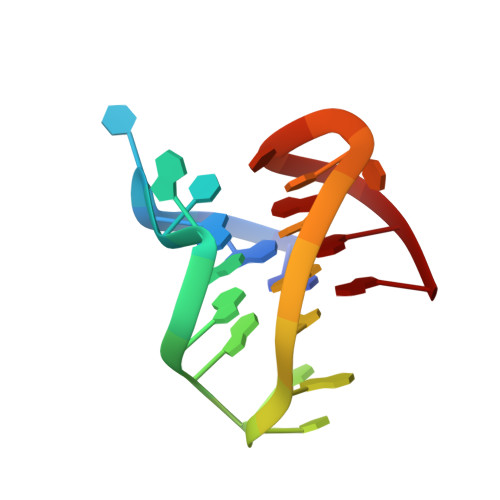
Ruthenium Polypyridyl Complex Bound to a Unimolecular Chair-Form G-Quadruplex.
McQuaid, K.T., Takahashi, S., Baumgaertner, L., Cardin, D.J., Paterson, N.G., Hall, J.P., Sugimoto, N., Cardin, C.J.(2022) J Am Chem Soc 144: 5956-5964
- PubMed: 35324198
- DOI: https://doi.org/10.1021/jacs.2c00178
- Primary Citation of Related Structures:
7OTB - PubMed Abstract:
The DNA G-quadruplex is known for forming a range of topologies and for the observed lability of the assembly, consistent with its transient formation in live cells. The stabilization of a particular topology by a small molecule is of great importance for therapeutic applications. Here, we show that the ruthenium complex Λ-[Ru(phen) 2 (qdppz)] 2+ displays enantiospecific G-quadruplex binding. It crystallized in 1:1 stoichiometry with a modified human telomeric G-quadruplex sequence, GGGTTAGGGTTAGGGTTTGGG ( htel21 T 18 ), in an antiparallel chair topology, the first structurally characterized example of ligand binding to this topology. The lambda complex is bound in an intercalation cavity created by a terminal G-quartet and the central narrow lateral loop formed by T 10 -T 11 -A 12 . The two remaining wide lateral loops are linked through a third K + ion at the other end of the G-quartet stack, which also coordinates three thymine residues. In a comparative ligand-binding study, we showed, using a Klenow fragment assay, that this complex is the strongest observed inhibitor of replication, both using the native human telomeric sequence and the modified sequence used in this work.
- Department of Chemistry, University of Reading, Whiteknights, Reading RG6 6AD, U.K.
Organizational Affiliation: