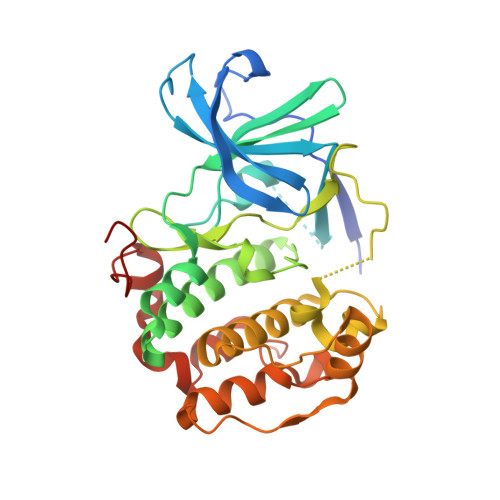
A non-catalytic herpesviral protein reconfigures ERK-RSK signaling by targeting kinase docking systems in the host.
Alexa, A., Sok, P., Gross, F., Albert, K., Kobori, E., Poti, A.L., Gogl, G., Bento, I., Kuang, E., Taylor, S.S., Zhu, F., Ciliberto, A., Remenyi, A.(2022) Nat Commun 13: 472-472
- PubMed: 35078976
- DOI: https://doi.org/10.1038/s41467-022-28109-x
- Primary Citation of Related Structures:
7OPM, 7OPO - PubMed Abstract:
The Kaposi's sarcoma associated herpesvirus protein ORF45 binds the extracellular signal-regulated kinase (ERK) and the p90 Ribosomal S6 kinase (RSK). ORF45 was shown to be a kinase activator in cells but a kinase inhibitor in vitro, and its effects on the ERK-RSK complex are unknown. Here, we demonstrate that ORF45 binds ERK and RSK using optimized linear binding motifs. The crystal structure of the ORF45-ERK2 complex shows how kinase docking motifs recognize the activated form of ERK. The crystal structure of the ORF45-RSK2 complex reveals an AGC kinase docking system, for which we provide evidence that it is functional in the host. We find that ORF45 manipulates ERK-RSK signaling by favoring the formation of a complex, in which activated kinases are better protected from phosphatases and docking motif-independent RSK substrate phosphorylation is selectively up-regulated. As such, our data suggest that ORF45 interferes with the natural design of kinase docking systems in the host.
- Biomolecular Interactions Research Group, Institute of Organic Chemistry, Research Center for Natural Sciences, H-1117, Budapest, Hungary.
Organizational Affiliation: