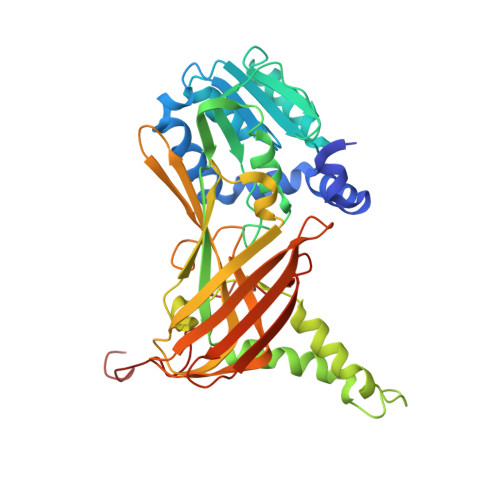
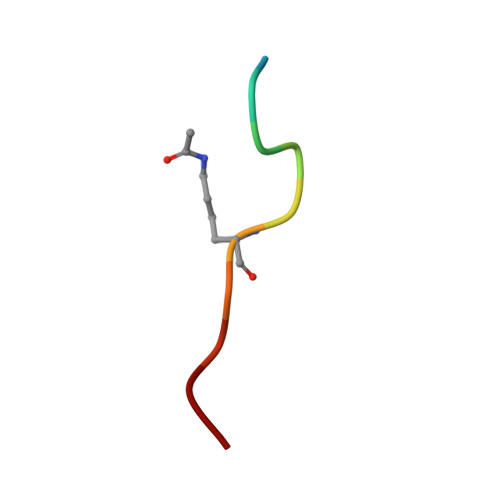
Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Zhang, Y., Marechal, N., van Haren, M.J., Troffer-Charlier, N., Cura, V., Cavarelli, J., Martin, N.I.(2021) Chembiochem 22: 3469-3476
- PubMed: 34569136
- DOI: https://doi.org/10.1002/cbic.202100506
- Primary Citation of Related Structures:
7OKP, 7OS4 - PubMed Abstract:
The dynamic interplay of post-translational modifications (PTMs) in chromatin provides a communication system for the regulation of gene expression. An increasing number of studies have highlighted the role that such crosstalk between PTMs plays in chromatin recognition. In this study, (bio)chemical and structural approaches were applied to specifically probe the impact of acetylation of Lys 18 in the histone H3 tail peptide on peptide recognition by the protein methyltransferase coactivator-associated arginine methyltransferase 1 (CARM1). Peptidomimetics that recapitulate the transition state of protein arginine N-methyltransferases, were designed based on the H3 peptide wherein the target Arg 17 was flanked by either a free or an acetylated lysine. Structural studies with these peptidomimetics and the catalytic domain of CARM1 provide new insights into the binding of the H3 peptide within the enzyme active site. While the co-crystal structures reveal that lysine acetylation results in minor conformational differences for both CARM1 and the H3 peptide, acetylation of Lys 18 does lead to additional interactions (Van der Waals and hydrogen bonding) and likely reduces the cost of desolvation upon binding, resulting in increased affinity. Informed by these findings a series of smaller peptidomimetics were also prepared and found to maintain potent and selective CARM1 inhibition. These findings provide new insights both into the mechanism of crosstalk between arginine methylation and lysine acetylation as well as towards the development of peptidomimetic CARM1 inhibitors.
- Biological Chemistry Group, Institute of Biology Leiden, Leiden University, Sylviusweg 72, 2333 BE, Leiden (The, Netherlands.
Organizational Affiliation: