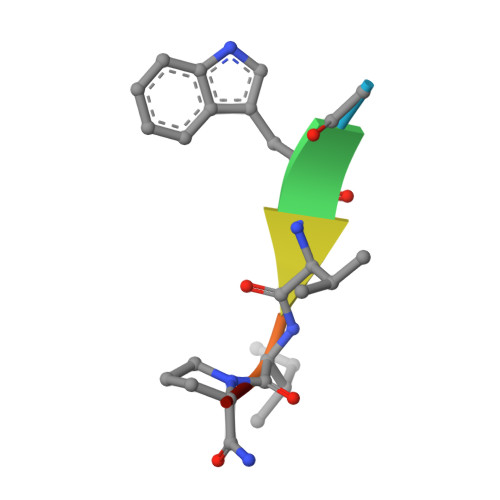
Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket.
Lloyd, M.G., Huckvale, R., Cheung, K.J., Rodrigues, M.J., Collie, G.W., Pierrat, O.A., Gatti Iou, M., Carter, M., Davis, O.A., McAndrew, P.C., Gunnell, E., Le Bihan, Y.V., Talbot, R., Henley, A.T., Johnson, L.D., Hayes, A., Bright, M.D., Raynaud, F.I., Meniconi, M., Burke, R., van Montfort, R.L.M., Rossanese, O.W., Bellenie, B.R., Hoelder, S.(2021) J Med Chem 64: 17079-17097
- PubMed: 34846884
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00946
- Primary Citation of Related Structures:
7OKD, 7OKE, 7OKF, 7OKG, 7OKH, 7OKI, 7OKJ, 7OKK, 7OKL, 7OKM - PubMed Abstract:
We describe the optimization of modestly active starting points to potent inhibitors of BCL6 by growing into a subpocket, which was occupied by a network of five stably bound water molecules. Identifying potent inhibitors required not only forming new interactions in the subpocket but also perturbing the water network in a productive, potency-increasing fashion while controlling the physicochemical properties. We achieved this goal in a sequential manner by systematically probing the pocket and the water network, ultimately achieving a 100-fold improvement of activity. The most potent compounds displaced three of the five initial water molecules and formed hydrogen bonds with the remaining two. Compound 25 showed a promising profile for a lead compound with submicromolar inhibition of BCL6 in cells and satisfactory pharmacokinetic (PK) properties. Our work highlights the importance of finding productive ways to perturb existing water networks when growing into solvent-filled protein pockets.