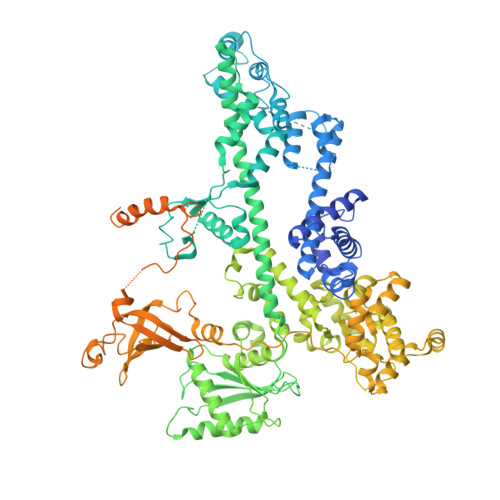
Cooperation between intrinsically disordered and ordered regions of Spt6 regulates nucleosome and Pol II CTD binding, and nucleosome assembly.
Kasiliauskaite, A., Kubicek, K., Klumpler, T., Zanova, M., Zapletal, D., Koutna, E., Novacek, J., Stefl, R.(2022) Nucleic Acids Res 50: 5961-5973
- PubMed: 35640611
- DOI: https://doi.org/10.1093/nar/gkac451
- Primary Citation of Related Structures:
7O3D, 7O6B - PubMed Abstract:
Transcription elongation factor Spt6 associates with RNA polymerase II (Pol II) and acts as a histone chaperone, which promotes the reassembly of nucleosomes following the passage of Pol II. The precise mechanism of nucleosome reassembly mediated by Spt6 remains unclear. In this study, we used a hybrid approach combining cryo-electron microscopy and small-angle X-ray scattering to visualize the architecture of Spt6 from Saccharomyces cerevisiae. The reconstructed overall architecture of Spt6 reveals not only the core of Spt6, but also its flexible N- and C-termini, which are critical for Spt6's function. We found that the acidic N-terminal region of Spt6 prevents the binding of Spt6 not only to the Pol II CTD and Pol II CTD-linker, but also to pre-formed intact nucleosomes and nucleosomal DNA. The N-terminal region of Spt6 self-associates with the tSH2 domain and the core of Spt6 and thus controls binding to Pol II and nucleosomes. Furthermore, we found that Spt6 promotes the assembly of nucleosomes in vitro. These data indicate that the cooperation between the intrinsically disordered and structured regions of Spt6 regulates nucleosome and Pol II CTD binding, and also nucleosome assembly.
- CEITEC-Central European Institute of Technology, Masaryk University, Brno CZ-62500, Czech Republic.
Organizational Affiliation: