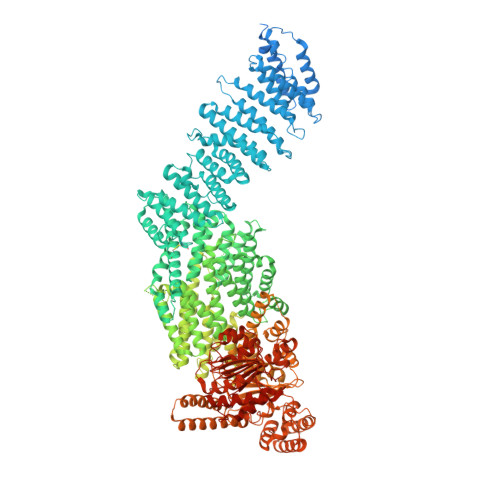
Structural basis of human separase regulation by securin and CDK1-cyclin B1.
Yu, J., Raia, P., Ghent, C.M., Raisch, T., Sadian, Y., Cavadini, S., Sabale, P.M., Barford, D., Raunser, S., Morgan, D.O., Boland, A.(2021) Nature 596: 138-142
- PubMed: 34290405
- DOI: https://doi.org/10.1038/s41586-021-03764-0
- Primary Citation of Related Structures:
7NJ0, 7NJ1 - PubMed Abstract:
In early mitosis, the duplicated chromosomes are held together by the ring-shaped cohesin complex 1 . Separation of chromosomes during anaphase is triggered by separase-a large cysteine endopeptidase that cleaves the cohesin subunit SCC1 (also known as RAD21 2-4 ). Separase is activated by degradation of its inhibitors, securin 5 and cyclin B 6 , but the molecular mechanisms of separase regulation are not clear. Here we used cryogenic electron microscopy to determine the structures of human separase in complex with either securin or CDK1-cyclin B1-CKS1. In both complexes, separase is inhibited by pseudosubstrate motifs that block substrate binding at the catalytic site and at nearby docking sites. As in Caenorhabditis elegans 7 and yeast 8 , human securin contains its own pseudosubstrate motifs. By contrast, CDK1-cyclin B1 inhibits separase by deploying pseudosubstrate motifs from intrinsically disordered loops in separase itself. One autoinhibitory loop is oriented by CDK1-cyclin B1 to block the catalytic sites of both separase and CDK1 9,10 . Another autoinhibitory loop blocks substrate docking in a cleft adjacent to the separase catalytic site. A third separase loop contains a phosphoserine 6 that promotes complex assembly by binding to a conserved phosphate-binding pocket in cyclin B1. Our study reveals the diverse array of mechanisms by which securin and CDK1-cyclin B1 bind and inhibit separase, providing the molecular basis for the robust control of chromosome segregation.
- Department of Molecular Biology, University of Geneva, Geneva, Switzerland.
Organizational Affiliation: