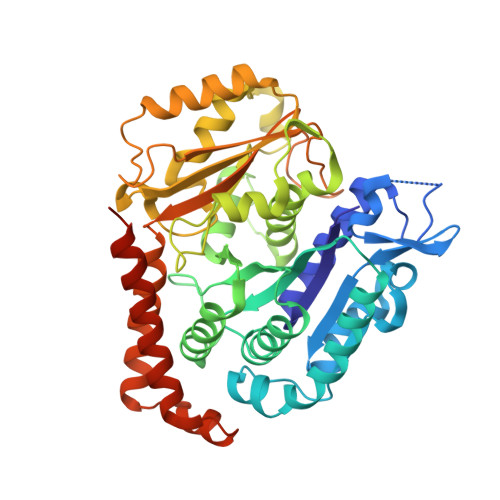
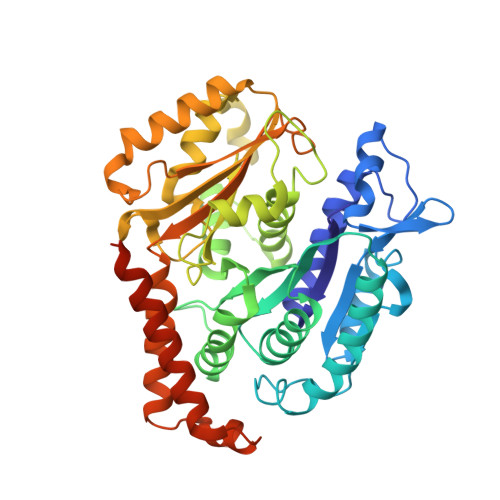
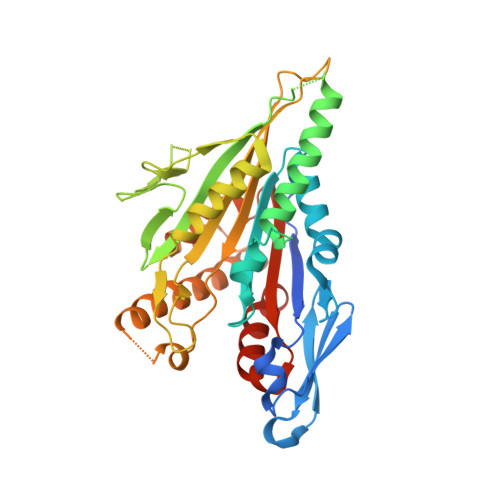
Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.
Cook, A.D., Roberts, A.J., Atherton, J., Tewari, R., Topf, M., Moores, C.A.(2021) J Biological Chem 297: 101063-101063
- PubMed: 34375637
- DOI: https://doi.org/10.1016/j.jbc.2021.101063
- Primary Citation of Related Structures:
7NB8, 7NBA - PubMed Abstract:
Plasmodium parasites cause malaria and are responsible annually for hundreds of thousands of deaths. Kinesins are a superfamily of microtubule-dependent ATPases that play important roles in the parasite replicative machinery, which is a potential target for antiparasite drugs. Kinesin-5, a molecular motor that cross-links microtubules, is an established antimitotic target in other disease contexts, but its mechanism in Plasmodium falciparum is unclear. Here, we characterized P. falciparum kinesin-5 (PfK5) using cryo-EM to determine the motor's nucleotide-dependent microtubule-bound structure and introduced 3D classification of individual motors into our microtubule image processing pipeline to maximize our structural insights. Despite sequence divergence in PfK5, the motor exhibits classical kinesin mechanochemistry, including ATP-induced subdomain rearrangement and cover neck bundle formation, consistent with its plus-ended directed motility. We also observed that an insertion in loop5 of the PfK5 motor domain creates a different environment in the well-characterized human kinesin-5 drug-binding site. Our data reveal the possibility for selective inhibition of PfK5 and can be used to inform future exploration of Plasmodium kinesins as antiparasite targets.
- Institute of Structural and Molecular Biology, Department of Biological Sciences, Birkbeck, University of London, London, United Kingdom.
Organizational Affiliation: