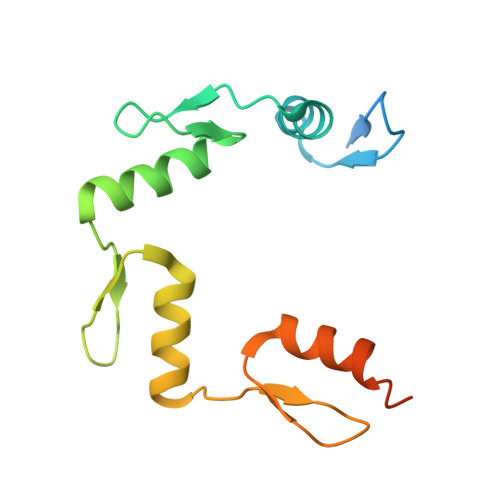
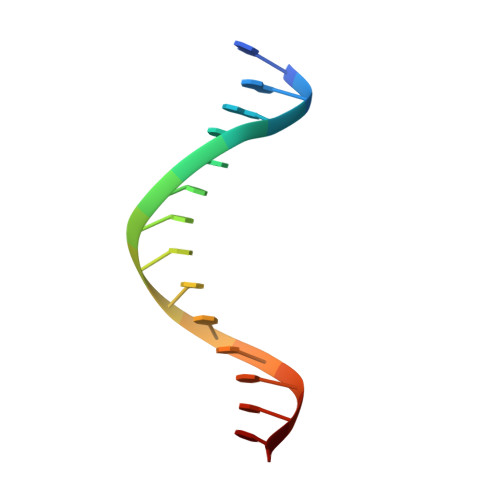
Structural basis for human ZBTB7A action at the fetal globin promoter.
Yang, Y., Ren, R., Ly, L.C., Horton, J.R., Li, F., Quinlan, K.G.R., Crossley, M., Shi, Y., Cheng, X.(2021) Cell Rep 36: 109759-109759
- PubMed: 34592153
- DOI: https://doi.org/10.1016/j.celrep.2021.109759
- Primary Citation of Related Structures:
7EYI, 7N5S, 7N5T - PubMed Abstract:
Elevated levels of fetal globin protect against β-hemoglobinopathies, such as sickle cell disease and β-thalassemia. Two zinc-finger (ZF) repressors, BCL11A and ZBTB7A/LRF, bind directly to the fetal globin promoter elements positioned at -115 and -200, respectively. Here, we describe X-ray structures of the ZBTB7A DNA-binding domain, consisting of four adjacent ZFs, in complex with the -200 sequence element, which contains two copies of four consecutive C:G base pairs. ZF1 and ZF2 recognize the 5' C:G quadruple, and ZF4 contacts the 3' C:G quadruple. Natural non-coding DNA mutations associated with hereditary persistence of fetal hemoglobin (HPFH) impair ZBTB7A DNA binding, with the most severe disruptions resulting from mutations in the base pairs recognized by ZF1 and ZF2. Our results firmly establish ZBTB7A/LRF as a key molecular regulator of fetal globin expression and inform genome-editing strategies that inhibit repressor binding and boost fetal globin expression to treat hemoglobinopathies.
- Hefei National Laboratory for Physical Sciences at Microscale, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui 230026, China; Ministry of Education Key Laboratory for Membraneless Organelles and Cellular Dynamics, University of Science and Technology of China, Hefei, China.
Organizational Affiliation: