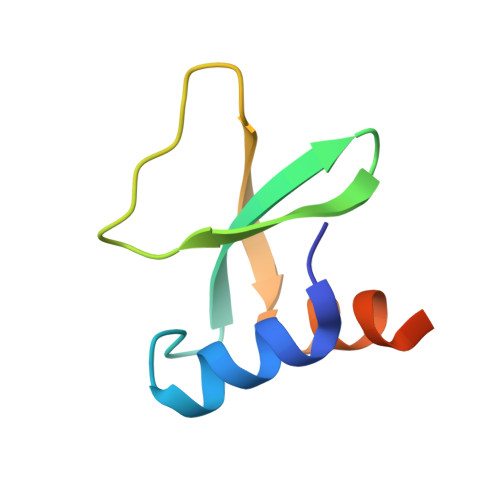
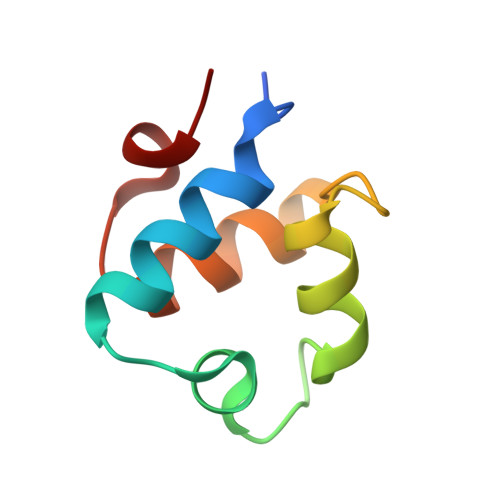
Molecular mechanism of quorum sensing inhibition in Streptococcus by the phage protein paratox.
Rutbeek, N.R., Rezasoltani, H., Patel, T.R., Khajehpour, M., Prehna, G.(2021) J Biological Chem 297: 100992-100992
- PubMed: 34298018
- DOI: https://doi.org/10.1016/j.jbc.2021.100992
- Primary Citation of Related Structures:
7N10, 7N1N - PubMed Abstract:
Streptococcus pyogenes, or Group A Streptococcus, is a Gram-positive bacterium that can be both a human commensal and a pathogen. Central to this dichotomy are temperate bacteriophages that incorporate into the bacterial genome as prophages. These genetic elements encode both the phage proteins and the toxins harmful to the human host. One such conserved phage protein, paratox (Prx), is always found encoded adjacent to the toxin genes, and this linkage is preserved during all stages of the phage life cycle. Within S. pyogenes, Prx functions to inhibit the quorum-sensing receptor-signal pair ComRS, the master regulator of natural competence, or the ability to uptake endogenous DNA. However, the mechanism by which Prx directly binds and inhibits the receptor ComR is unknown. To understand how Prx inhibits ComR at the molecular level, we pursued an X-ray crystal structure of Prx bound to ComR. The structural data supported by solution X-ray scattering data demonstrate that Prx induces a conformational change in ComR to directly access its DNA-binding domain. Furthermore, electromobility shift assays and competition binding assays reveal that Prx effectively uncouples the interdomain conformational change required for activation of ComR via the signaling molecule XIP. Although to our knowledge the molecular mechanism of quorum-sensing inhibition by Prx is unique, it is analogous to the mechanism employed by the phage protein Aqs1 in Pseudomonas aeruginosa. Together, this demonstrates an example of convergent evolution between Gram-positive and Gram-negative phages to inhibit quorum-sensing and highlights the versatility of small phage proteins.
- Department of Microbiology, University of Manitoba, Winnipeg, Manitoba, Canada.
Organizational Affiliation: