CryoEM and AI reveal a structure of SARS-CoV-2 Nsp2, a multifunctional protein involved in key host processes.
QCRG Structural Biology ConsortiumTo be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Non-structural protein 2 | 638 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b | 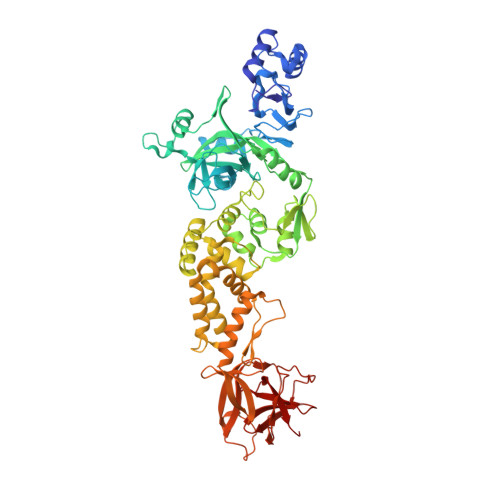 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN Query on ZN | B [auth A], C [auth A], D [auth A] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | Rosetta | 2020.08.61146 |
| MODEL REFINEMENT | ISOLDE | 1.0 |
| RECONSTRUCTION | cryoSPARC | v2.15.0 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| DARPA | United States | HR0011-19-2-0020 |
| FastGrants | United States | COVID19 grant |
| Laboratory for Genomics Research | United States | Excellence in Research Award COVID19 |
| Quantitative Biosciences Institute | United States | -- |