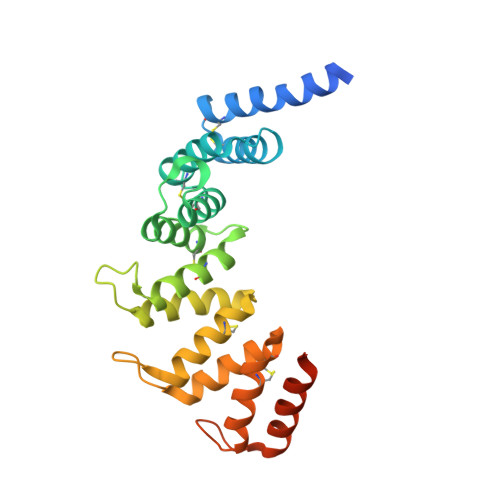
Mitochondrial COA7 is a heme-binding protein with disulfide reductase activity, which acts in the early stages of complex IV assembly.
Formosa, L.E., Maghool, S., Sharpe, A.J., Reljic, B., Muellner-Wong, L., Stroud, D.A., Ryan, M.T., Maher, M.J.(2022) Proc Natl Acad Sci U S A 119
- PubMed: 35210360
- DOI: https://doi.org/10.1073/pnas.2110357119
- Primary Citation of Related Structures:
7MQZ - PubMed Abstract:
Cytochrome c oxidase (COX) assembly factor 7 (COA7) is a metazoan-specific assembly factor, critical for the biogenesis of mitochondrial complex IV (cytochrome c oxidase). Although mutations in COA7 have been linked to complex IV assembly defects and neurological conditions such as peripheral neuropathy, ataxia, and leukoencephalopathy, the precise role COA7 plays in the biogenesis of complex IV is not known. Here, we show that loss of COA7 blocks complex IV assembly after the initial step where the COX1 module is built, progression from which requires the incorporation of copper and addition of the COX2 and COX3 modules. The crystal structure of COA7, determined to 2.4 Å resolution, reveals a banana-shaped molecule composed of five helix-turn-helix (α/α) repeats, tethered by disulfide bonds. COA7 interacts transiently with the copper metallochaperones SCO1 and SCO2 and catalyzes the reduction of disulfide bonds within these proteins, which are crucial for copper relay to COX2. COA7 binds heme with micromolar affinity, through axial ligation to the central iron atom by histidine and methionine residues. We therefore propose that COA7 is a heme-binding disulfide reductase for regenerating the copper relay system that underpins complex IV assembly.
- Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, VIC 3800, Australia.
Organizational Affiliation: