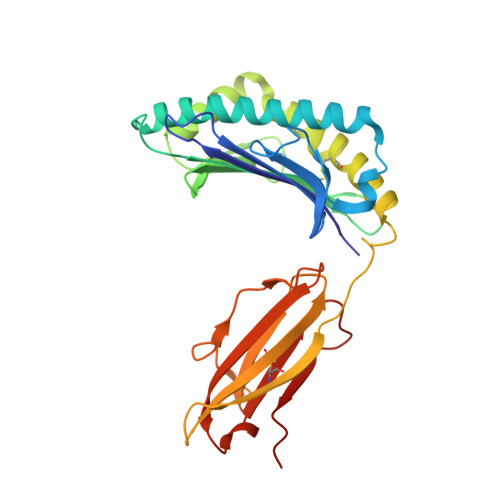
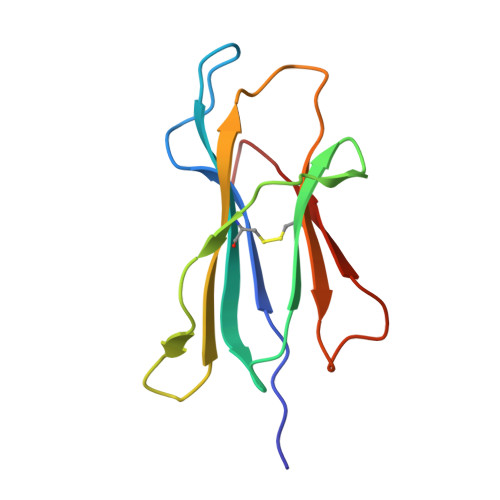
Homologous peptides derived from influenza A, B and C viruses induce variable CD8 + T cell responses with cross-reactive potential.
Nguyen, A.T., Lau, H.M.P., Sloane, H., Jayasinghe, D., Mifsud, N.A., Chatzileontiadou, D.S., Grant, E.J., Szeto, C., Gras, S.(2022) Clin Transl Immunology 11: e1422-e1422
- PubMed: 36275878
- DOI: https://doi.org/10.1002/cti2.1422
- Primary Citation of Related Structures:
7MLE - PubMed Abstract:
Influenza A, B and C viruses (IAV, IBV and ICV, respectively) circulate globally, infecting humans and causing widespread morbidity and mortality. Here, we investigate the T cell response towards an immunodominant IAV epitope, NP 265-273, and its IBV and ICV homologues, presented by HLA-A*03:01 molecule expressed in ~ 4% of the global population (~ 300 million people). We assessed the magnitude (tetramer staining) and quality of the CD8 + T cell response (intracellular cytokine staining) towards NP 265-IAV and described the T cell receptor (TCR) repertoire used to recognise this immunodominant epitope. We next assessed the immunogenicity of NP 265-IAV homologue peptides from IBV and ICV and the ability of CD8 + T cells to cross-react towards these homologous peptides. Furthermore, we determined the structures of NP 265-IAV and NP 323-IBV peptides in complex with HLA-A*03:01 by X-ray crystallography. Our study provides a detailed characterisation of the CD8 + T cell response towards NP 265-IAV and its IBV and ICV homologues. The data revealed a diverse repertoire for NP 265-IAV that is associated with superior anti-viral protection. Evidence of cross-reactivity between the three different influenza virus strain-derived epitopes was observed, indicating the discovery of a potential vaccination target that is broad enough to cover all three influenza strains. We show that while there is a potential to cross-protect against distinct influenza virus lineages, the T cell response was stronger against the IAV peptide than IBV or ICV, which is an important consideration when choosing targets for future vaccine design.
- Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute Monash University Clayton VIC Australia.
Organizational Affiliation: