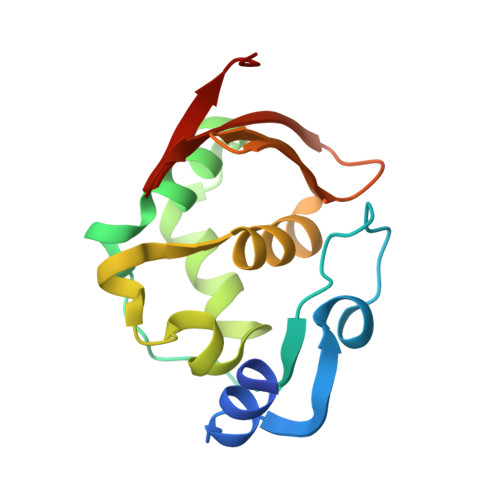
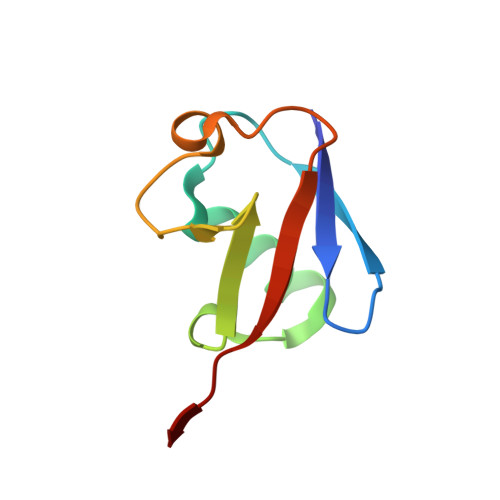
The endopeptidase of the maize-affecting Marafivirus type member maize rayado fino virus doubles as a deubiquitinase.
Patel, A., McBride, J.A.M., Mark, B.L.(2021) J Biological Chem 297: 100957-100957
- PubMed: 34265303
- DOI: https://doi.org/10.1016/j.jbc.2021.100957
- Primary Citation of Related Structures:
7MIA, 7MIC - PubMed Abstract:
Marafiviruses are capable of persistent infection in a range of plants that have importance to the agriculture and biofuel industries. Although the genomes of a few of these viruses have been studied in-depth, the composition and processing of the polyproteins produced from their main ORFs have not. The Marafivirus polyprotein consists of essential proteins that form the viral replicase, as well as structural proteins for virus assembly. It has been proposed that Marafiviruses code for cysteine proteases within their polyproteins, which act as endopeptidases to autocatalytically cleave the polyprotein into functional domains. Furthermore, it has also been suggested that Marafivirus endopeptidases may have deubiquitinating activity, which has been shown to enhance viral replication by downregulating viral protein degradation by the ubiquitin (Ub) proteasomal pathway as well as tampering with cell signaling associated with innate antiviral responses in other positive-sense ssRNA viruses. Here, we provide the first evidence of cysteine proteases from six different Marafiviruses that harbor deubiquitinating activity and reveal intragenus differences toward Ub linkage types. We also examine the structural basis of the endopeptidase/deubiquitinase from the Marafivirus type member, maize rayado fino virus. Structures of the enzyme alone and bound to Ub reveal marked structural rearrangements that occur upon binding of Ub and provide insights into substrate specificity and differences that set it apart from other viral cysteine proteases.
- Department of Microbiology, University of Manitoba, Winnipeg, Canada.
Organizational Affiliation: