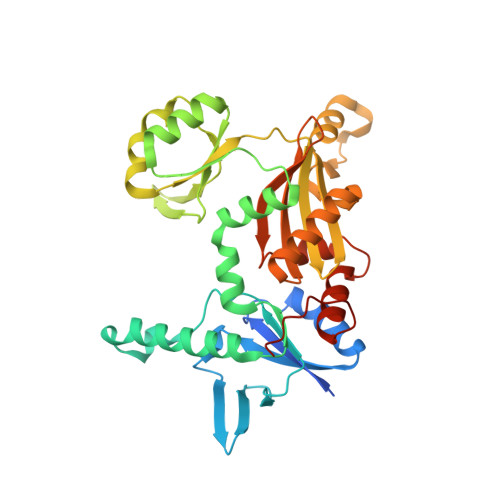
Structural Basis for a Dual Function ATP Grasp Ligase That Installs Single and Bicyclic omega-Ester Macrocycles in a New Multicore RiPP Natural Product.
Zhao, G., Kosek, D., Liu, H.B., Ohlemacher, S.I., Blackburne, B., Nikolskaya, A., Makarova, K.S., Sun, J., Barry Iii, C.E., Koonin, E.V., Dyda, F., Bewley, C.A.(2021) J Am Chem Soc 143: 8056-8068
- PubMed: 34028251
- DOI: https://doi.org/10.1021/jacs.1c02316
- Primary Citation of Related Structures:
7MGV - PubMed Abstract:
Among the ribosomally synthesized and post-translationally modified peptide (RiPP) natural products, "graspetides" (formerly known as microviridins) contain macrocyclic esters and amides that are formed by ATP-grasp ligase tailoring enzymes using the side chains of Asp/Glu as acceptors and Thr/Ser/Lys as donors. Graspetides exhibit diverse patterns of macrocylization and connectivities exemplified by microviridins, that have a caged tricyclic core, and thuringin and plesiocin that feature a "hairpin topology" with cross-strand ω-ester bonds. Here, we characterize chryseoviridin, a new type of multicore RiPP encoded by Chryseobacterium gregarium DS19109 (Phylum Bacteroidetes) and solve a 2.44 Å resolution crystal structure of a quaternary complex consisting of the ATP-grasp ligase CdnC bound to ADP, a conserved leader peptide and a peptide substrate. HRMS/MS analyses show that chryseoviridin contains four consecutive five- or six-residue macrocycles ending with a microviridin-like core. The crystal structure captures respective subunits of the CdnC homodimer in the apo or substrate-bound state revealing a large conformational change in the B-domain upon substrate binding. A docked model of ATP places the γ-phosphate group within 2.8 Å of the Asp acceptor residue. The orientation of the bound substrate is consistent with a model in which macrocyclization occurs in the N- to C-terminal direction for core peptides containing multiple Thr/Ser-to-Asp macrocycles. Using systematically varied sequences, we validate this model and identify two- or three-amino acid templating elements that flank the macrolactone and are required for enzyme activity in vitro. This work reveals the structural basis for ω-ester bond formation in RiPP biosynthesis.
- Laboratory of Bioorganic Chemistry, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892, United States.
Organizational Affiliation: