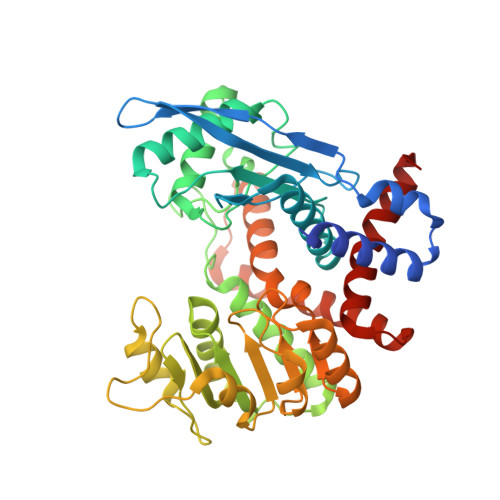
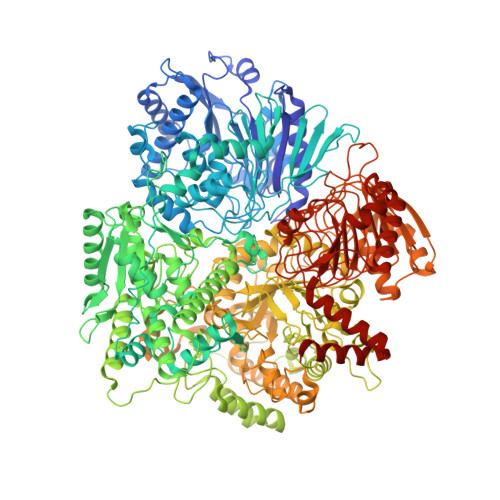
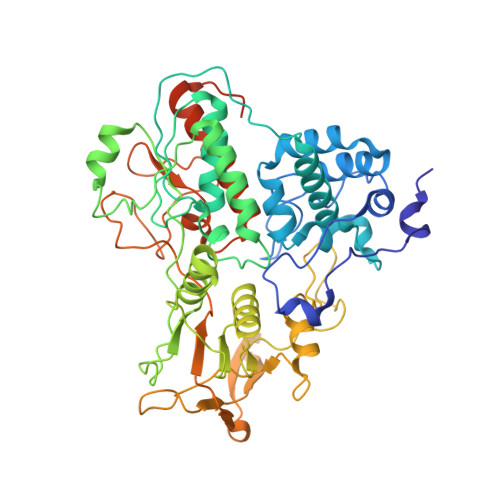
A counter-enzyme complex regulates glutamate metabolism in Bacillus subtilis.
Jayaraman, V., Lee, D.J., Elad, N., Vimer, S., Sharon, M., Fraser, J.S., Tawfik, D.S.(2022) Nat Chem Biol 18: 161-170
- PubMed: 34931064
- DOI: https://doi.org/10.1038/s41589-021-00919-y
- Primary Citation of Related Structures:
7MFM, 7MFT - PubMed Abstract:
Multi-enzyme assemblies composed of metabolic enzymes catalyzing sequential reactions are being increasingly studied. Here, we report the discovery of a 1.6 megadalton multi-enzyme complex from Bacillus subtilis composed of two enzymes catalyzing opposite ('counter-enzymes') rather than sequential reactions: glutamate synthase (GltAB) and glutamate dehydrogenase (GudB), which make and break glutamate, respectively. In vivo and in vitro studies show that the primary role of complex formation is to inhibit the activity of GudB. Using cryo-electron microscopy, we elucidated the structure of the complex and the molecular basis of inhibition of GudB by GltAB. The complex exhibits unusual oscillatory progress curves and is necessary for both planktonic growth, in glutamate-limiting conditions, and for biofilm growth, in glutamate-rich media. The regulation of a key metabolic enzyme by complexing with its counter enzyme may thus enable cell growth under fluctuating glutamate concentrations.
- Department of Biomolecular Sciences, Weizmann Institute of Science, Rehovot, Israel. vijay.jayaraman@weizmann.ac.il.
Organizational Affiliation: