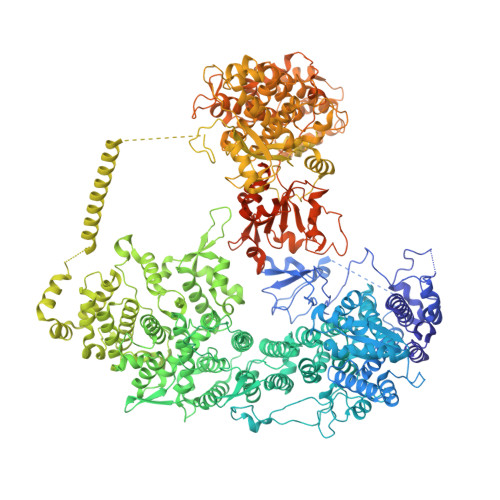
Structural insights into Ubr1-mediated N-degron polyubiquitination.
Pan, M., Zheng, Q., Wang, T., Liang, L., Mao, J., Zuo, C., Ding, R., Ai, H., Xie, Y., Si, D., Yu, Y., Liu, L., Zhao, M.(2021) Nature 600: 334-338
- PubMed: 34789879
- DOI: https://doi.org/10.1038/s41586-021-04097-8
- Primary Citation of Related Structures:
7MEX, 7MEY - PubMed Abstract:
The N-degron pathway targets proteins that bear a destabilizing residue at the N terminus for proteasome-dependent degradation 1 . In yeast, Ubr1-a single-subunit E3 ligase-is responsible for the Arg/N-degron pathway 2 . How Ubr1 mediates the initiation of ubiquitination and the elongation of the ubiquitin chain in a linkage-specific manner through a single E2 ubiquitin-conjugating enzyme (Ubc2) remains unknown. Here we developed chemical strategies to mimic the reaction intermediates of the first and second ubiquitin transfer steps, and determined the cryo-electron microscopy structures of Ubr1 in complex with Ubc2, ubiquitin and two N-degron peptides, representing the initiation and elongation steps of ubiquitination. Key structural elements, including a Ubc2-binding region and an acceptor ubiquitin-binding loop on Ubr1, were identified and characterized. These structures provide mechanistic insights into the initiation and elongation of ubiquitination catalysed by Ubr1.
- Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL, USA. panman@sjtu.edu.cn.
Organizational Affiliation: