Comprehensive structural survey of DNA double-strand break synapsis by DNA Polymerase Lambda
Kaminski, A.M., Bebenek, K., Kunkel, T.A., Pedersen, L.C.Not Published
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Not Published
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA polymerase lambda | 346 | Homo sapiens | Mutation(s): 0 Gene Names: POLL EC: 2.7.7.7 (PDB Primary Data), 4.2.99 (PDB Primary Data) | 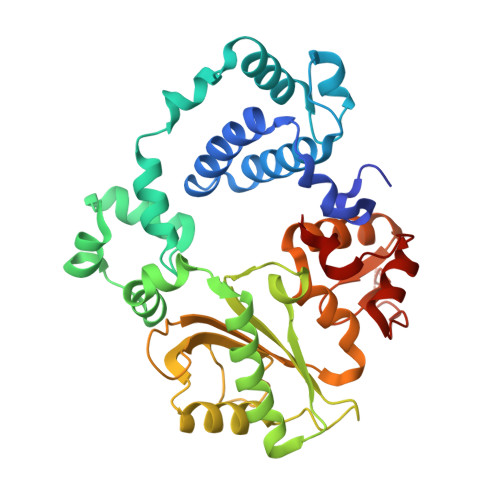 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9UGP5 (Homo sapiens) Explore Q9UGP5 Go to UniProtKB: Q9UGP5 | |||||
PHAROS: Q9UGP5 GTEx: ENSG00000166169 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9UGP5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*CP*GP*GP*CP*AP*G)-3') | B [auth T] | 6 | synthetic construct | 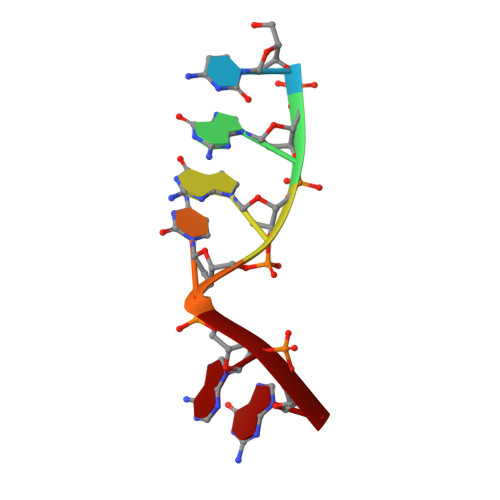 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*TP*AP*CP*TP*G)-3') | C [auth U] | 5 | synthetic construct | 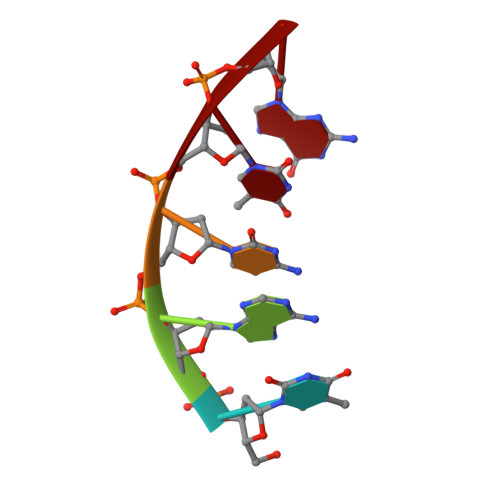 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*CP*AP*GP*TP*GP*CP*T)-3') | D [auth P] | 7 | synthetic construct | 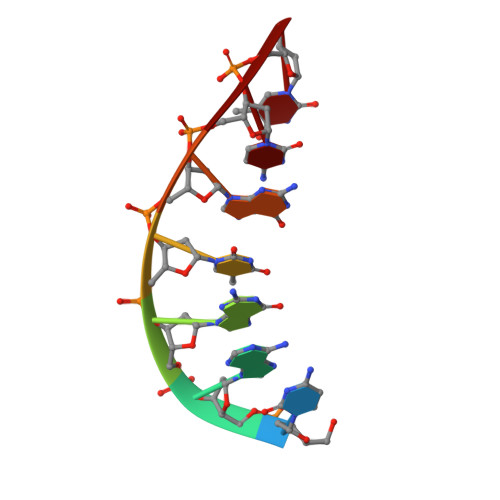 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(P*GP*CP*CP*G)-3') | E [auth D] | 4 | synthetic construct |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PPV Query on PPV | L [auth A] | PYROPHOSPHATE H4 O7 P2 XPPKVPWEQAFLFU-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | M [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| EDO Query on EDO | N [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CL Query on CL | J [auth A], K [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | F [auth A], G [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| NA Query on NA | H [auth A], I [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 56.205 | α = 90 |
| b = 60.023 | β = 90 |
| c = 139.824 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of Environmental Health Sciences (NIH/NIEHS) | United States | 1ZIA ES102645 |
| National Institutes of Health/National Institute of Environmental Health Sciences (NIH/NIEHS) | United States | Z01 ES065070 |