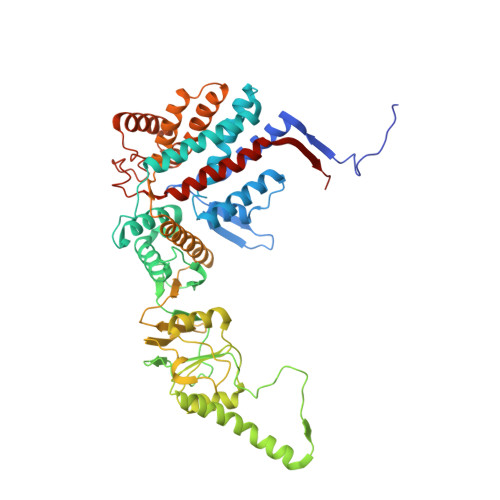
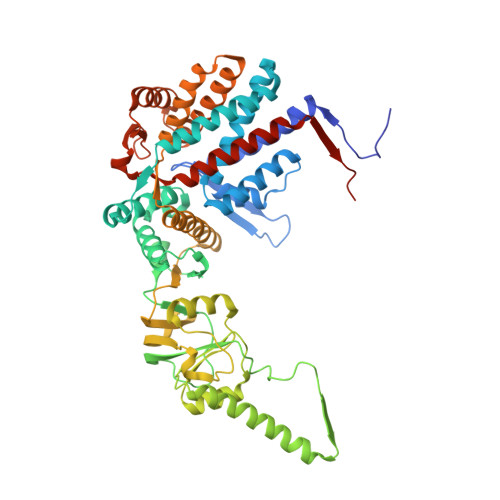
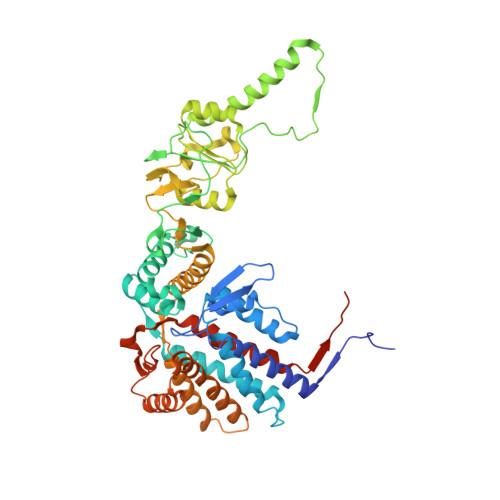
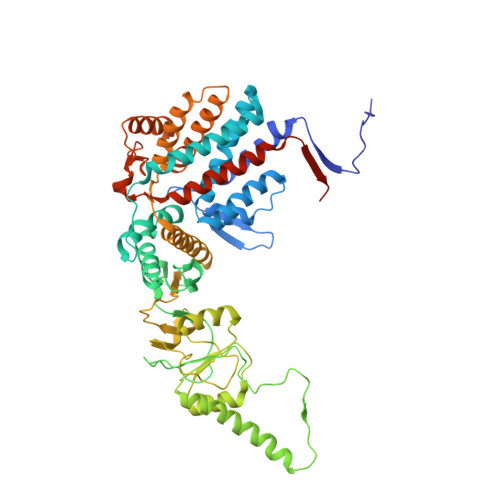
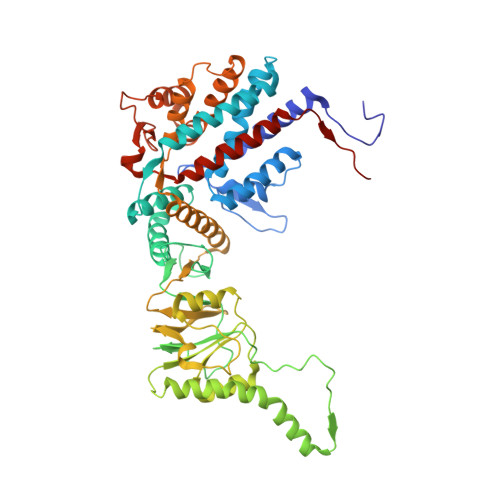
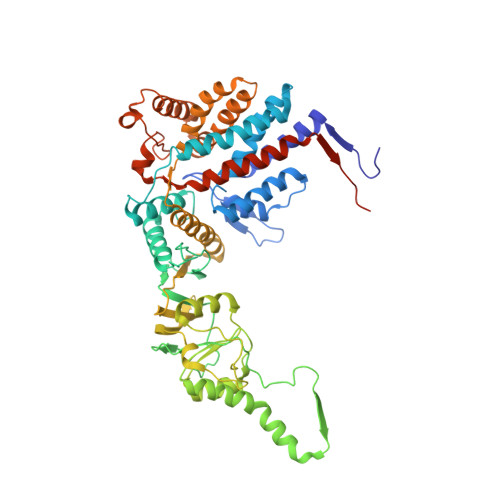
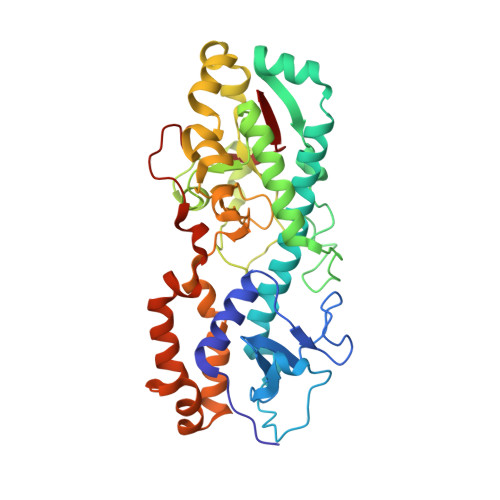
Structural and functional dissection of reovirus capsid folding and assembly by the prefoldin-TRiC/CCT chaperone network.
Knowlton, J.J., Gestaut, D., Ma, B., Taylor, G., Seven, A.B., Leitner, A., Wilson, G.J., Shanker, S., Yates, N.A., Prasad, B.V.V., Aebersold, R., Chiu, W., Frydman, J., Dermody, T.S.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33836586
- DOI: https://doi.org/10.1073/pnas.2018127118
- Primary Citation of Related Structures:
7LUM, 7LUP - PubMed Abstract:
Intracellular protein homeostasis is maintained by a network of chaperones that function to fold proteins into their native conformation. The eukaryotic TRiC chaperonin (TCP1-ring complex, also called CCT for cytosolic chaperonin containing TCP1) facilitates folding of a subset of proteins with folding constraints such as complex topologies. To better understand the mechanism of TRiC folding, we investigated the biogenesis of an obligate TRiC substrate, the reovirus σ3 capsid protein. We discovered that the σ3 protein interacts with a network of chaperones, including TRiC and prefoldin. Using a combination of cryoelectron microscopy, cross-linking mass spectrometry, and biochemical approaches, we establish functions for TRiC and prefoldin in folding σ3 and promoting its assembly into higher-order oligomers. These studies illuminate the molecular dynamics of σ3 folding and establish a biological function for TRiC in virus assembly. In addition, our findings provide structural and functional insight into the mechanism by which TRiC and prefoldin participate in the assembly of protein complexes.
- Department of Pediatrics, University of Pittsburgh School of Medicine, Pittsburgh, PA 15224.
Organizational Affiliation: