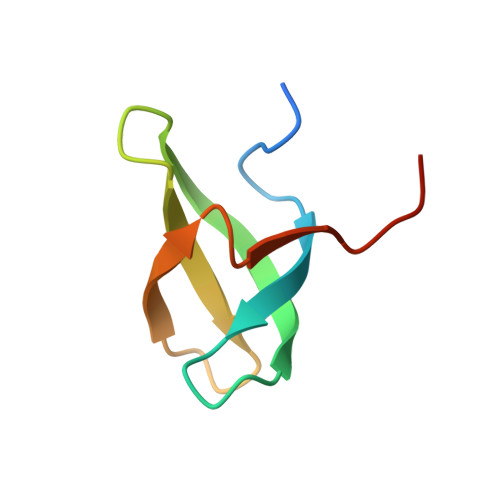
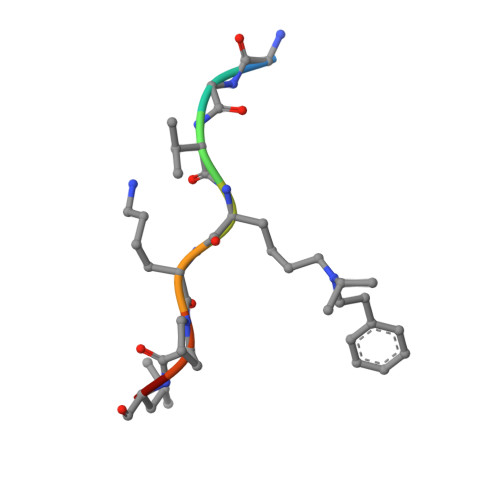
Discovery of an H3K36me3-Derived Peptidomimetic Ligand with Enhanced Affinity for Plant Homeodomain Finger Protein 1 (PHF1).
Engelberg, I.A., Liu, J., Norris-Drouin, J.L., Cholensky, S.H., Ottavi, S.A., Frye, S.V., Kutateladze, T.G., James, L.I.(2021) J Med Chem 64: 8510-8522
- PubMed: 33999620
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00430
- Primary Citation of Related Structures:
7LKY - PubMed Abstract:
Plant homeodomain finger protein 1 (PHF1) is an accessory component of the gene silencing complex polycomb repressive complex 2 and recognizes the active chromatin mark, trimethylated lysine 36 of histone H3 (H3K36me3). In addition to its role in transcriptional regulation, PHF1 has been implicated as a driver of endometrial stromal sarcoma and fibromyxoid tumors. We report the discovery and characterization of UNC6641, a peptidomimetic antagonist of the PHF1 Tudor domain which was optimized through in silico modeling and incorporation of non-natural amino acids. UNC6641 binds the PHF1 Tudor domain with a K d value of 0.96 ± 0.03 μM while also binding the related protein PHF19 with similar potency. A crystal structure of PHF1 in complex with UNC6641, along with NMR and site-directed mutagenesis data, provided insight into the binding mechanism and requirements for binding. Additionally, UNC6641 enabled the development of a high-throughput assay to identify small molecule binders of PHF1.
- Center for Integrative Chemical Biology and Drug Discovery, Division of Chemical Biology and Medicinal Chemistry, UNC Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599, United States.
Organizational Affiliation: