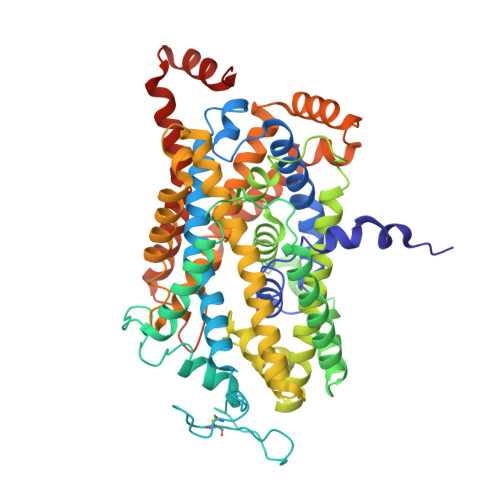
Illumination of serotonin transporter mechanism and role of the allosteric site.
Yang, D., Gouaux, E.(2021) Sci Adv 7: eabl3857-eabl3857
- PubMed: 34851672
- DOI: https://doi.org/10.1126/sciadv.abl3857
- Primary Citation of Related Structures:
7LI6, 7LI7, 7LI8, 7LI9, 7LIA, 7MGW - PubMed Abstract:
The serotonin transporter (SERT) terminates serotonin signaling by using sodium and chloride gradients to drive reuptake of serotonin into presynaptic neurons and is the target of widely used medications to treat neuropsychiatric disorders. Despite decades of study, the molecular mechanism of serotonin transport, the coupling to ion gradients, and the role of the allosteric site have remained elusive. Here, we present cryo–electron microscopy structures of SERT in serotonin-bound and serotonin-free states, in the presence of sodium or potassium, resolving all fundamental states of the transport cycle. From the SERT-serotonin complex, we localize the substrate-bound allosteric site, formed by an aromatic pocket positioned in the scaffold domain in the extracellular vestibule, connected to the central site via a short tunnel. Together with elucidation of multiple apo state conformations, we provide previously unseen structural understanding of allosteric modulation, demonstrating how SERT binds serotonin from synaptic volumes and promotes unbinding into the presynaptic neurons.
- Vollum Institute, Oregon Health and Science University, Portland, OR 97239, USA.
Organizational Affiliation: