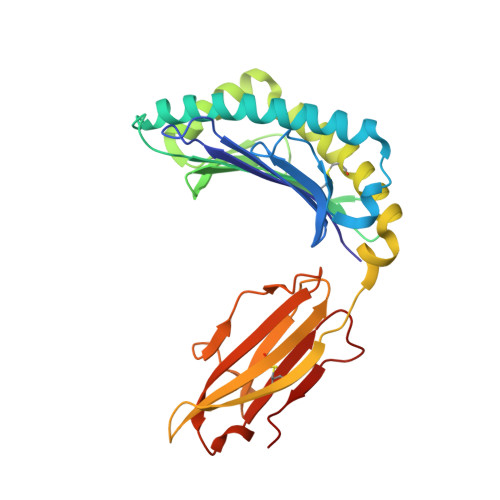
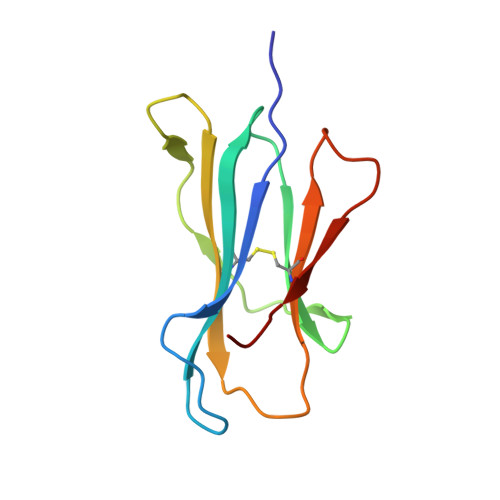
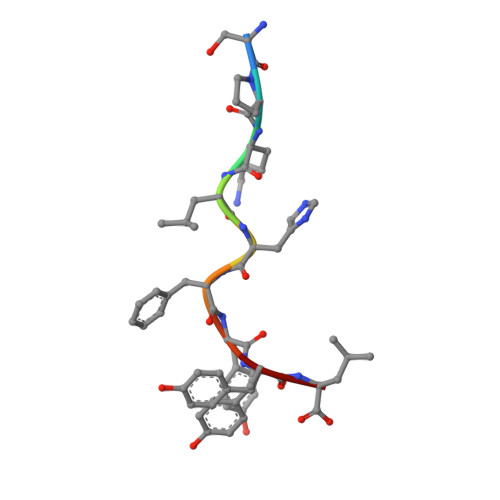
CD8 + T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses.
Lineburg, K.E., Grant, E.J., Swaminathan, S., Chatzileontiadou, D.S.M., Szeto, C., Sloane, H., Panikkar, A., Raju, J., Crooks, P., Rehan, S., Nguyen, A.T., Lekieffre, L., Neller, M.A., Tong, Z.W.M., Jayasinghe, D., Chew, K.Y., Lobos, C.A., Halim, H., Burrows, J.M., Riboldi-Tunnicliffe, A., Chen, W., D'Orsogna, L., Khanna, R., Short, K.R., Smith, C., Gras, S.(2021) Immunity 54: 1055
- PubMed: 33945786
- DOI: https://doi.org/10.1016/j.immuni.2021.04.006
- Primary Citation of Related Structures:
7LGD, 7LGT - PubMed Abstract:
Efforts are being made worldwide to understand the immune response to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for the coronavirus disease 2019 (COVID-19) pandemic, including the impact of T cell immunity and cross-recognition with seasonal coronaviruses. Screening of SARS-CoV-2 peptide pools revealed that the nucleocapsid (N) protein induced an immunodominant response in HLA-B7 + COVID-19-recovered individuals that was also detectable in unexposed donors. A single N-encoded epitope that was highly conserved across circulating coronaviruses drove this immunodominant response. In vitro peptide stimulation and crystal structure analyses revealed T cell-mediated cross-reactivity toward circulating OC43 and HKU-1 betacoronaviruses but not 229E or NL63 alphacoronaviruses because of different peptide conformations. T cell receptor (TCR) sequencing indicated that cross-reactivity was driven by private TCR repertoires with a bias for TRBV27 and a long CDR3β loop. Our findings demonstrate the basis of selective T cell cross-reactivity for an immunodominant SARS-CoV-2 epitope and its homologs from seasonal coronaviruses, suggesting long-lasting protective immunity.
- QIMR Berghofer Centre for Immunotherapy and Vaccine Development and Translational and Human Immunology Laboratory, Department of Immunology, QIMR Berghofer Medical Research Institute, Brisbane, QLD 4006, Australia.
Organizational Affiliation: