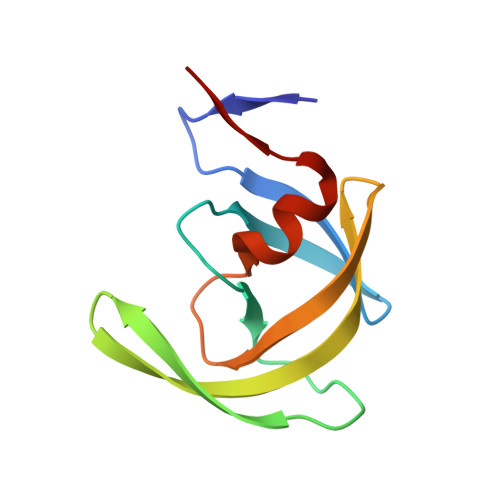
HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
Lockbaum, G.J., Rusere, L.N., Henes, M., Kosovrasti, K., Lee, S.K., Spielvogel, E., Nalivaika, E.A., Swanstrom, R., KurtYilmaz, N., Schiffer, C.A., Ali, A.To be published.