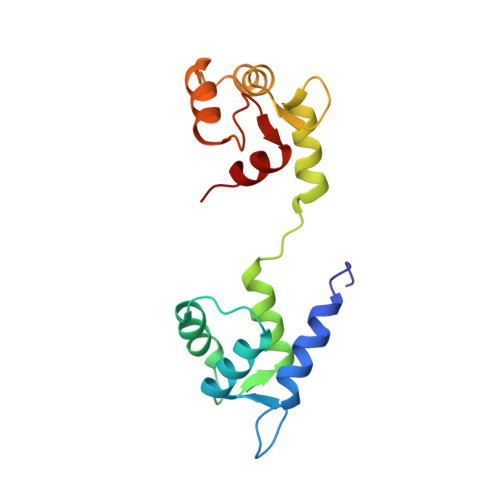
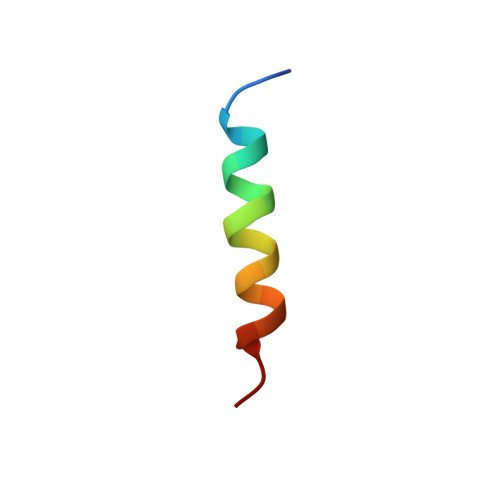
Half-calcified calmodulin promotes basal activity and inactivation of the L-type calcium channel Ca V 1.2.
Bartels, P., Salveson, I., Coleman, A.M., Anderson, D.E., Jeng, G., Estrada-Tobar, Z.M., Man, K.N.M., Yu, Q., Kuzmenkina, E., Nieves-Cintron, M., Navedo, M.F., Horne, M.C., Hell, J.W., Ames, J.B.(2022) J Biological Chem 298: 102701-102701
- PubMed: 36395884
- DOI: https://doi.org/10.1016/j.jbc.2022.102701
- Primary Citation of Related Structures:
7L8V - PubMed Abstract:
The L-type Ca 2+ channel Ca V 1.2 controls gene expression, cardiac contraction, and neuronal activity. Calmodulin (CaM) governs Ca V 1.2 open probability (Po) and Ca 2+ -dependent inactivation (CDI) but the mechanisms remain unclear. Here, we present electrophysiological data that identify a half Ca 2+ -saturated CaM species (Ca 2 /CaM) with Ca 2+ bound solely at the third and fourth EF-hands (EF3 and EF4) under resting Ca 2+ concentrations (50-100 nM) that constitutively preassociates with Ca V 1.2 to promote Po and CDI. We also present an NMR structure of a complex between the Ca V 1.2 IQ motif (residues 1644-1665) and Ca 2 /CaM 12' , a calmodulin mutant in which Ca 2+ binding to EF1 and EF2 is completely disabled. We found that the CaM 12' N-lobe does not interact with the IQ motif. The CaM 12' C-lobe bound two Ca 2+ ions and formed close contacts with IQ residues I1654 and Y1657. I1654A and Y1657D mutations impaired CaM binding, CDI, and Po, as did disabling Ca 2+ binding to EF3 and EF4 in the CaM 34 mutant when compared to WT CaM. Accordingly, a previously unappreciated Ca 2 /CaM species promotes Ca V 1.2 Po and CDI, identifying Ca 2 /CaM as an important mediator of Ca signaling.
- Department of Pharmacology, University of California, Davis, California, USA.
Organizational Affiliation: