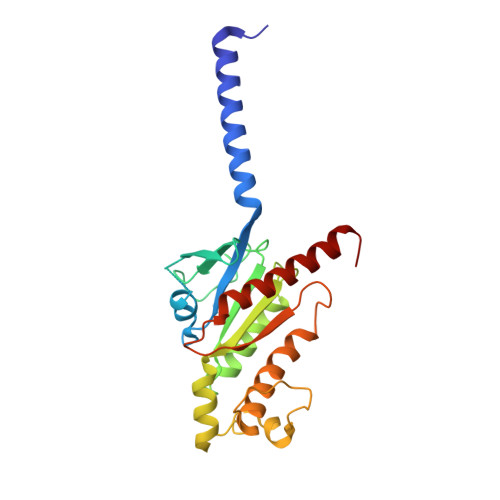
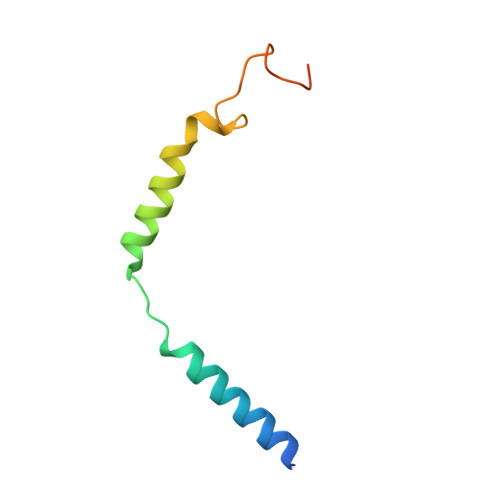
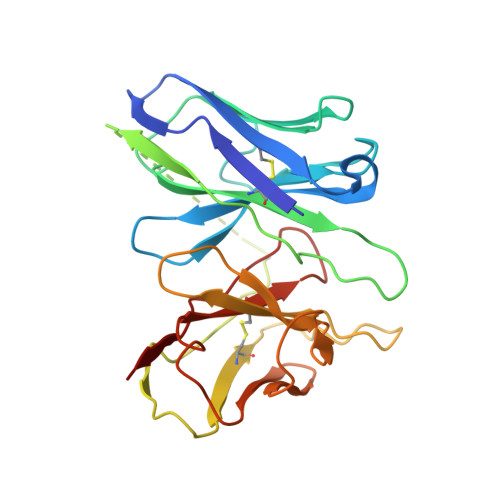
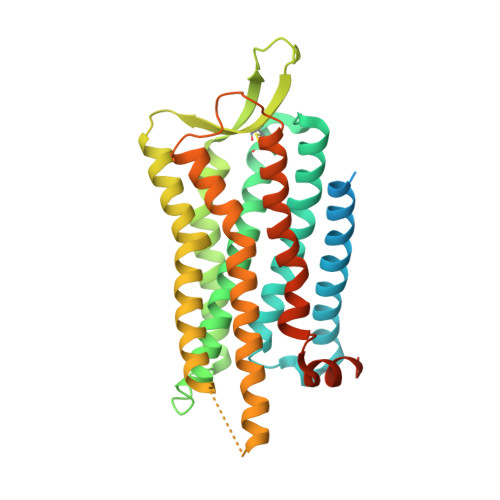
Structures of active-state orexin receptor 2 rationalize peptide and small-molecule agonist recognition and receptor activation.
Hong, C., Byrne, N.J., Zamlynny, B., Tummala, S., Xiao, L., Shipman, J.M., Partridge, A.T., Minnick, C., Breslin, M.J., Rudd, M.T., Stachel, S.J., Rada, V.L., Kern, J.C., Armacost, K.A., Hollingsworth, S.A., O'Brien, J.A., Hall, D.L., McDonald, T.P., Strickland, C., Brooun, A., Soisson, S.M., Hollenstein, K.(2021) Nat Commun 12: 815-815
- PubMed: 33547286
- DOI: https://doi.org/10.1038/s41467-021-21087-6
- Primary Citation of Related Structures:
7L1U, 7L1V - PubMed Abstract:
Narcolepsy type 1 (NT1) is a chronic neurological disorder that impairs the brain's ability to control sleep-wake cycles. Current therapies are limited to the management of symptoms with modest effectiveness and substantial adverse effects. Agonists of the orexin receptor 2 (OX 2 R) have shown promise as novel therapeutics that directly target the pathophysiology of the disease. However, identification of drug-like OX 2 R agonists has proven difficult. Here we report cryo-electron microscopy structures of active-state OX 2 R bound to an endogenous peptide agonist and a small-molecule agonist. The extended carboxy-terminal segment of the peptide reaches into the core of OX 2 R to stabilize an active conformation, while the small-molecule agonist binds deep inside the orthosteric pocket, making similar key interactions. Comparison with antagonist-bound OX 2 R suggests a molecular mechanism that rationalizes both receptor activation and inhibition. Our results enable structure-based discovery of therapeutic orexin agonists for the treatment of NT1 and other hypersomnia disorders.
- Computational & Structural Chemistry, MRL, Merck & Co., Inc, Kenilworth, NJ, USA.
Organizational Affiliation: