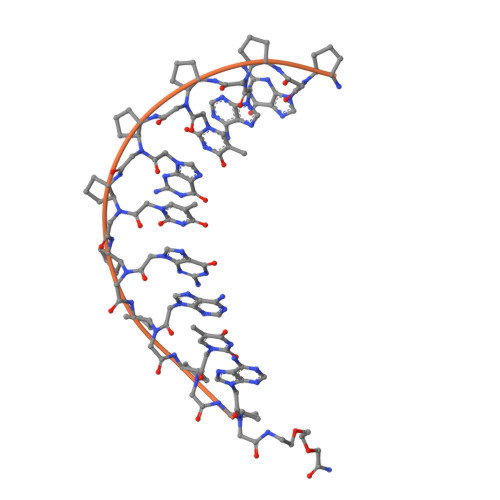
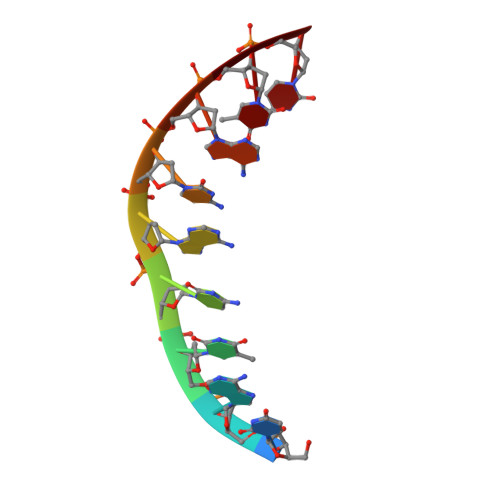
Conformational constraints of cyclopentane peptide nucleic acids facilitate tunable binding to DNA.
Zheng, H., Botos, I., Clausse, V., Nikolayevskiy, H., Rastede, E.E., Fouz, M.F., Mazur, S.J., Appella, D.H.(2021) Nucleic Acids Res 49: 713-725
- PubMed: 33406227
- DOI: https://doi.org/10.1093/nar/gkaa1249
- Primary Citation of Related Structures:
7KZL - PubMed Abstract:
We report a series of synthetic, nucleic acid mimics with highly customizable thermodynamic binding to DNA. Incorporation of helix-promoting cyclopentanes into peptide nucleic acids (PNAs) increases the melting temperatures (Tm) of PNA+DNA duplexes by approximately +5°C per cyclopentane. Sequential addition of cyclopentanes allows the Tm of PNA + DNA duplexes to be systematically fine-tuned from +5 to +50°C compared with the unmodified PNA. Containing only nine nucleobases and an equal number of cyclopentanes, cpPNA-9 binds to complementary DNA with a Tm around 90°C. Additional experiments reveal that the cpPNA-9 sequence specifically binds to DNA duplexes containing its complementary sequence and functions as a PCR clamp. An X-ray crystal structure of the cpPNA-9-DNA duplex revealed that cyclopentanes likely induce a right-handed helix in the PNA with conformations that promote DNA binding.
- Synthetic Bioactive Molecules Section, Laboratory of Bioorganic Chemistry (LBC), National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 8 Center Drive, Room 404, Bethesda, MD 20892, USA.
Organizational Affiliation: