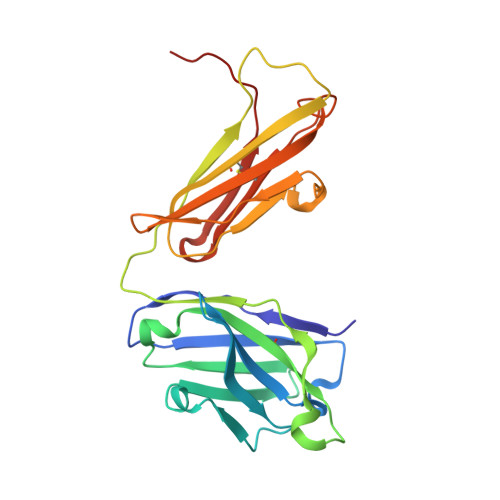
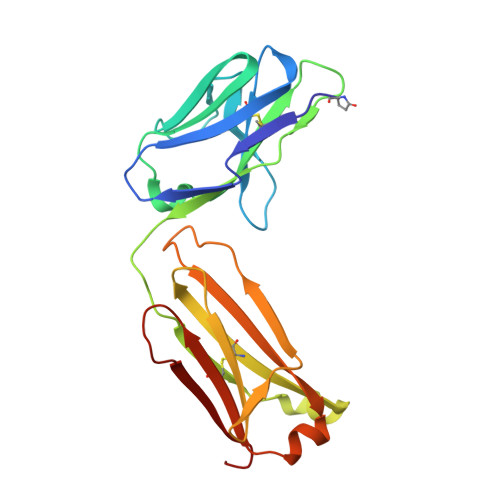
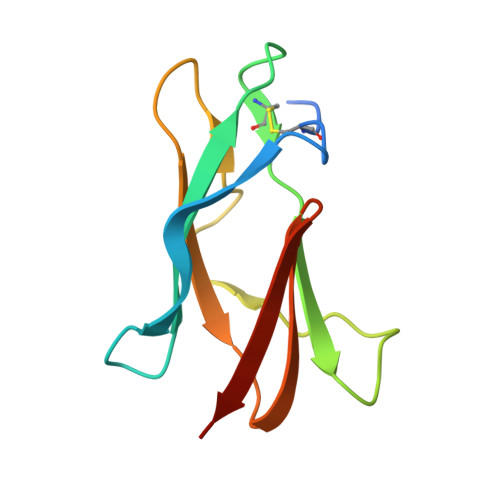
Broadly neutralizing monoclonal antibodies protect against multiple tick-borne flaviviruses.
VanBlargan, L.A., Errico, J.M., Kafai, N.M., Burgomaster, K.E., Jethva, P.N., Broeckel, R.M., Meade-White, K., Nelson, C.A., Himansu, S., Wang, D., Handley, S.A., Gross, M.L., Best, S.M., Pierson, T.C., Fremont, D.H., Diamond, M.S.(2021) J Exp Medicine 218
- PubMed: 33831142
- DOI: https://doi.org/10.1084/jem.20210174
- Primary Citation of Related Structures:
7KYL - PubMed Abstract:
Although Powassan virus (POWV) is an emerging tick-transmitted flavivirus that causes severe or fatal neuroinvasive disease in humans, medical countermeasures have not yet been developed. Here, we developed a panel of neutralizing anti-POWV mAbs recognizing six distinct antigenic sites. The most potent of these mAbs bind sites within domain II or III of the envelope (E) protein and inhibit postattachment viral entry steps. A subset of these mAbs cross-react with other flaviviruses. Both POWV type-specific and cross-reactive neutralizing mAbs confer protection in mice against POWV infection when given as prophylaxis or postexposure therapy. Several cross-reactive mAbs mapping to either domain II or III also protect in vivo against heterologous tick-transmitted flaviviruses including Langat and tick-borne encephalitis virus. Our experiments define structural and functional correlates of antibody protection against POWV infection and identify epitopes targeted by broadly neutralizing antibodies with therapeutic potential against multiple tick-borne flaviviruses.
- Department of Medicine, Washington University School of Medicine, St. Louis, MO.
Organizational Affiliation: