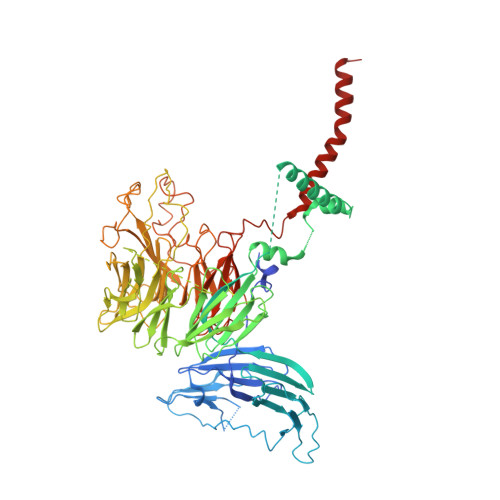
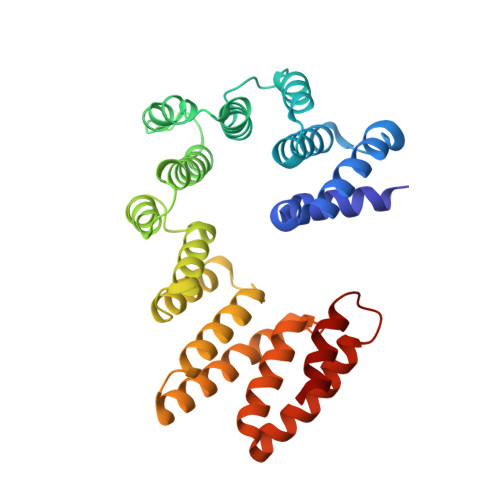
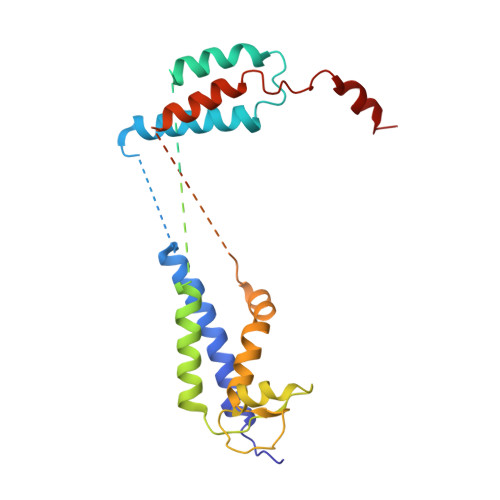
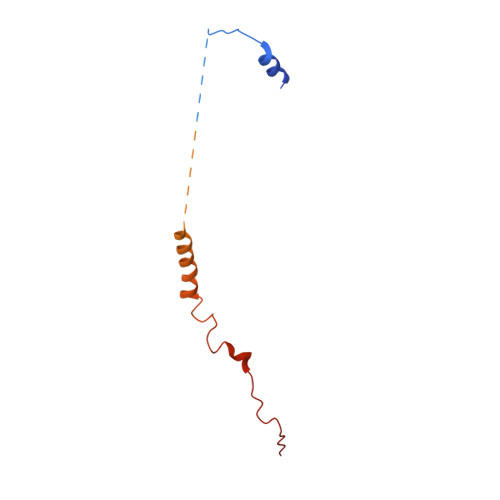
Structural and mechanistic basis of the EMC-dependent biogenesis of distinct transmembrane clients.
Miller-Vedam, L.E., Brauning, B., Popova, K.D., Schirle Oakdale, N.T., Bonnar, J.L., Prabu, J.R., Boydston, E.A., Sevillano, N., Shurtleff, M.J., Stroud, R.M., Craik, C.S., Schulman, B.A., Frost, A., Weissman, J.S.(2020) Elife 9
- PubMed: 33236988
- DOI: https://doi.org/10.7554/eLife.62611
- Primary Citation of Related Structures:
7ADO, 7ADP, 7KRA, 7KTX - PubMed Abstract:
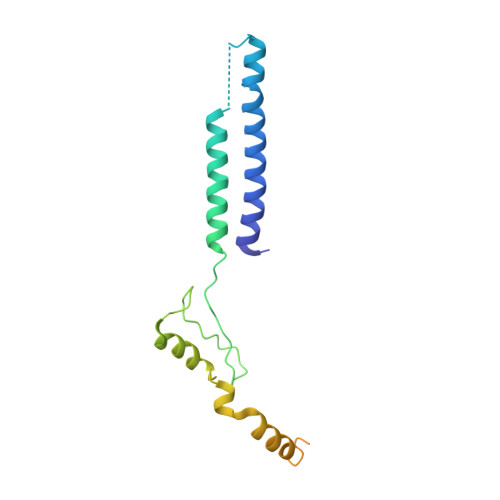
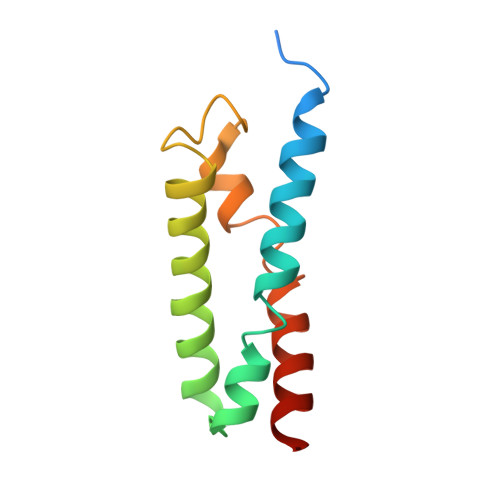
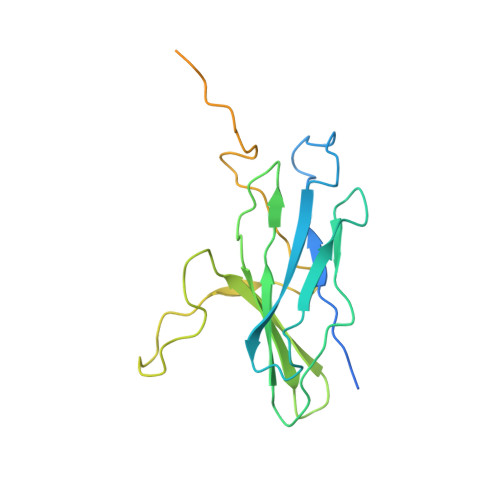
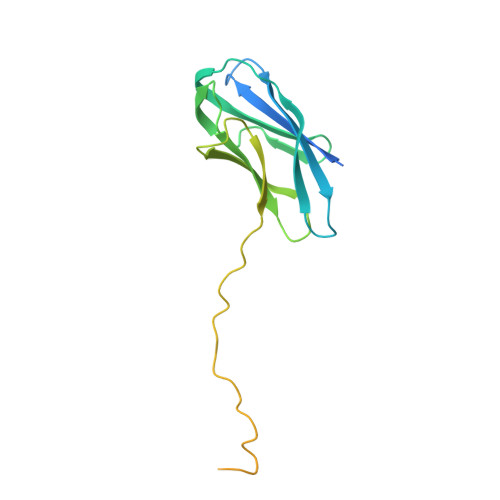
Membrane protein biogenesis in the endoplasmic reticulum (ER) is complex and failure-prone. The ER membrane protein complex (EMC), comprising eight conserved subunits, has emerged as a central player in this process. Yet, we have limited understanding of how EMC enables insertion and integrity of diverse clients, from tail-anchored to polytopic transmembrane proteins. Here, yeast and human EMC cryo-EM structures reveal conserved intricate assemblies and human-specific features associated with pathologies. Structure-based functional studies distinguish between two separable EMC activities, as an insertase regulating tail-anchored protein levels and a broader role in polytopic membrane protein biogenesis. These depend on mechanistically coupled yet spatially distinct regions including two lipid-accessible membrane cavities which confer client-specific regulation, and a non-insertase EMC function mediated by the EMC lumenal domain. Our studies illuminate the structural and mechanistic basis of EMC's multifunctionality and point to its role in differentially regulating the biogenesis of distinct client protein classes.
- Molecular, Cellular, and Computational Biophysics Graduate Program, University of California, San Francisco, San Francisco, United States.
Organizational Affiliation: