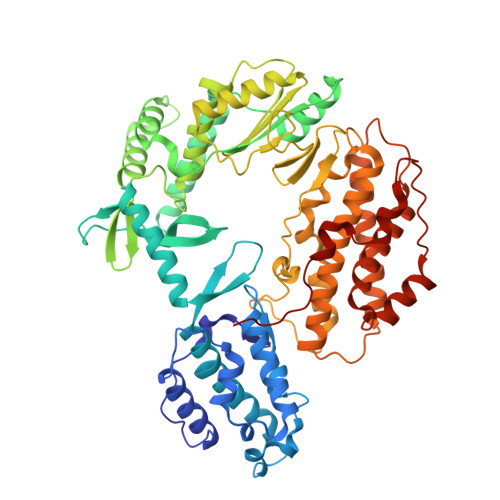
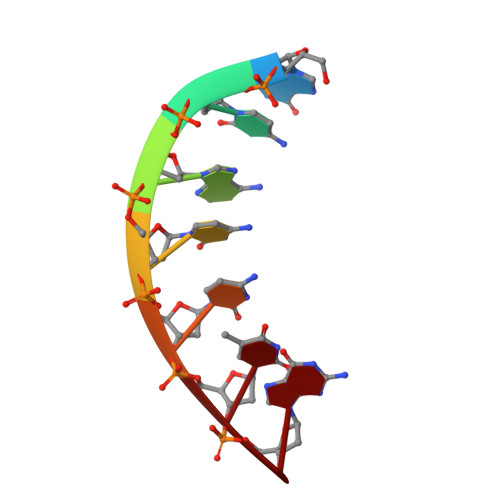
Flexibility of telomerase in binding the RNA template and DNA telomeric repeat.
Choi, W.S., Weng, P.J., Yang, W.(2022) Proc Natl Acad Sci U S A 119
- PubMed: 34969861
- DOI: https://doi.org/10.1073/pnas.2116159118
- Primary Citation of Related Structures:
7KQM, 7KQN - PubMed Abstract:
Telomerase synthesizes telomeres at the ends of linear chromosomes by repeated reverse transcription from a short RNA template. Crystal structures of Tribolium castaneum telomerase reverse transcriptase ( tc TERT) and cryoelectron microscopy (cryo-EM) structures of human and Tetrahymena telomerase have revealed conserved features in the reverse-transcriptase domain, including a cavity near the DNA 3' end and snug interactions with the RNA template. For the RNA template to translocate, it needs to be unpaired and separated from the DNA product. Here we investigate the potential of the structural cavity to accommodate a looped-out DNA bulge and enable the separation of the RNA/DNA hybrid. Using tc TERT as a model system, we show that a looped-out telomeric repeat in the DNA primer can be accommodated and extended by tc TERT but not by retroviral reverse transcriptase. Mutations that reduce the cavity size reduce the ability of tc TERT to extend the looped-out DNA substrate. In agreement with cryo-EM structures of telomerases, we find that tc TERT requires a minimum of 4 bp between the RNA template and DNA primer for efficient DNA synthesis. We also have determined the ternary-complex structure of tc TERT including a downstream RNA/DNA hybrid at 2.0-Å resolution and shown that a downstream RNA duplex, equivalent to the 5' template-boundary element in telomerase RNA, enhances the efficiency of telomere synthesis by tc TERT. Although TERT has a preformed active site without the open-and-closed conformational changes, it contains cavities to accommodate looped-out RNA and DNA. The flexible RNA-DNA binding likely underlies the processivity of telomeric repeat addition.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, NIH, Bethesda, MD 20892.
Organizational Affiliation: