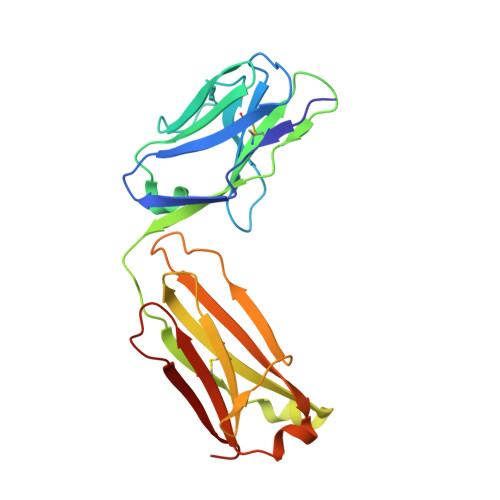
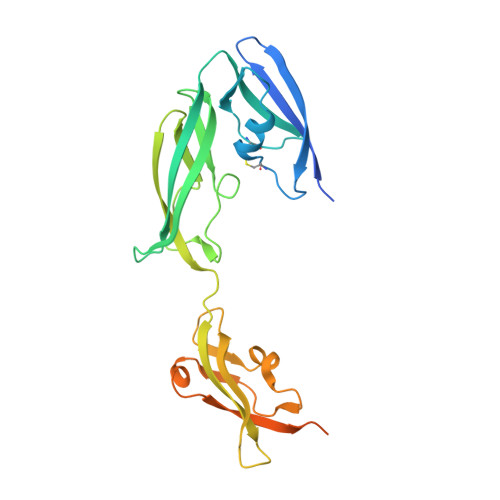
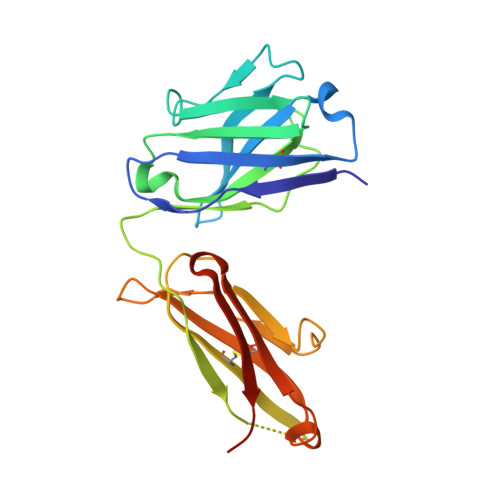
The molecular characterization of antibody binding to a superantigen-like protein from a commensal microbe.
Borowska, M.T., Drees, C., Yarawsky, A.E., Viswanathan, M., Ryan, S.M., Bunker, J.J., Herr, A.B., Bendelac, A., Adams, E.J.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34548394
- DOI: https://doi.org/10.1073/pnas.2023898118
- Primary Citation of Related Structures:
7KPJ - PubMed Abstract:
Microorganisms have coevolved diverse mechanisms to impair host defenses. A major one, superantigens, can result in devastating effects on the immune system. While all known superantigens induce vast immune cell proliferation and come from opportunistic pathogens, recently, proteins with similar broad specificity to antibody variable (V) domain families were identified in a commensal microbiota. These proteins, identified in the human commensal Ruminococcus gnavus , are called immunoglobulin-binding protein (Ibp) A and B and have been shown to activate B cells in vitro expressing either human VH3 or murine VH5/6/7. Here, we provide molecular and functional studies revealing the basis of this Ibp/immunoglobulin (Ig) interaction. The crystal structure and biochemical assays of a truncated IbpA construct in complex with mouse VH5 antigen-binding fragment (Fab) shows a binding of Ig heavy chain framework residues to the Ibp Domain D and the C-terminal heavy chain binding domain (HCBD). We used targeted mutagenesis of contact residues and affinity measurements and performed studies of the Fab-IbpA complex to determine the stoichiometry between Ibp and VH domains, suggesting Ibp may serve to cluster full-length IgA antibodies in vivo. Furthermore, in vitro stimulation experiments indicate that binding of the Ibp HCBD alone is sufficient to activate responsive murine B cell receptors. The presence of these proteins in a commensal microbe suggest that binding a broad repertoire of immunoglobulins, particularly in the gut/microbiome environment, may provide an important function in the maintenance of host/microbiome homeostasis contrasting with the pathogenic role of structurally homologous superantigens expressed by pathogens.
- Department of Biochemistry and Molecular Biology, University of Chicago, Chicago, IL 60637.
Organizational Affiliation: