Fluorescence-based Assay Development for Screening Novel Inhibitors of Dihydrodipicolinate Synthase from Campylobacter jejuni
Majdi Yazd, M., Reddy Annadi, K., Saran, S., Bhagwa, A., Sanders, D.A.R., Palmer, D.R.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 4-hydroxy-tetrahydrodipicolinate synthase | 310 | Campylobacter jejuni subsp. jejuni NCTC 11168 = ATCC 700819 | Mutation(s): 1 Gene Names: dapA, Cj0806 EC: 4.3.3.7 | 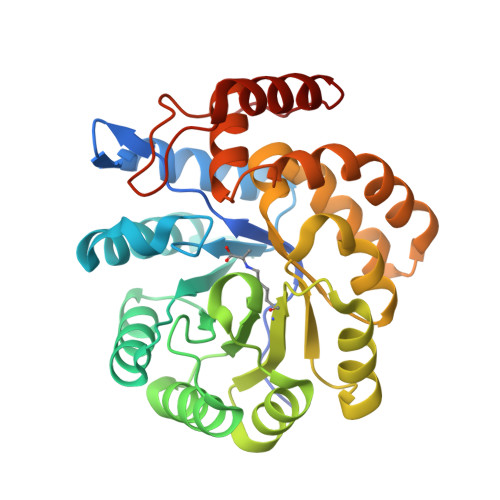 | |
UniProt | |||||
Find proteins for Q9PPB4 (Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168)) Explore Q9PPB4 Go to UniProtKB: Q9PPB4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9PPB4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PG4 Query on PG4 | BB [auth D], R [auth A] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| PGE Query on PGE | CB [auth E], FA [auth C], S [auth B], T [auth B] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | NA [auth C] O [auth A] OA [auth C] P [auth A] PA [auth C] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | EA [auth B], LB [auth E], MB [auth E], SA [auth C] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | AA [auth B] BA [auth B] CA [auth B] DA [auth B] FB [auth E] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | AB [auth D] KB [auth E] Q [auth A] QA [auth C] RA [auth C] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| MG Query on MG | DB [auth E] EB [auth E] G [auth A] GA [auth C] H [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| KPI Query on KPI | A, B, C, D, E A, B, C, D, E, F | L-PEPTIDE LINKING | C9 H16 N2 O4 |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 85.13 | α = 90 |
| b = 231.25 | β = 90 |
| c = 202.1 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XSCALE | data scaling |
| PDB_EXTRACT | data extraction |
| HKL-3000 | data reduction |
| AMPLE | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Natural Sciences and Engineering Research Council (NSERC, Canada) | Canada | -- |