Rational Design of RNA Editing Guide Strands: Cytidine Analogs at the Orphan Position.
Doherty, E.E., Wilcox, X.E., van Sint Fiet, L., Kemmel, C., Turunen, J.J., Klein, B., Tantillo, D.J., Fisher, A.J., Beal, P.A.(2021) J Am Chem Soc 143: 6865-6876
- PubMed: 33939417
- DOI: https://doi.org/10.1021/jacs.0c13319
- Primary Citation of Related Structures:
7KFN - PubMed Abstract:
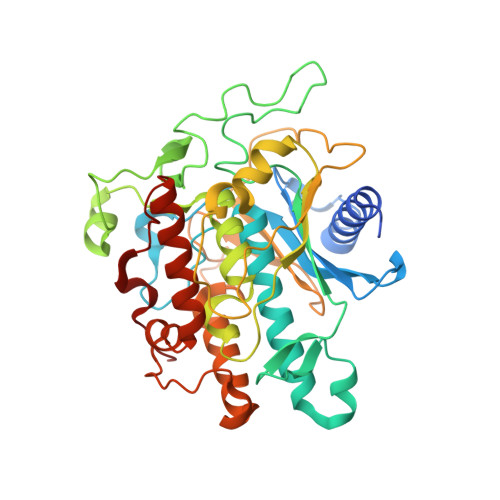
Adenosine Deaminases Acting on RNA (ADARs) convert adenosine to inosine in double stranded RNA. Human ADARs can be directed to predetermined target sites in the transcriptome by complementary guide strands, allowing for the correction of disease-causing mutations at the RNA level. Here we use structural information available for ADAR2-RNA complexes to guide the design of nucleoside analogs for the position in the guide strand that contacts a conserved glutamic acid residue in ADARs (E488 in human ADAR2), which flips the adenosine into the ADAR active site for deamination. Mutating this residue to glutamine (E488Q) results in higher activity because of the hydrogen bond donating ability of Q488 to N3 of the orphan cytidine on the guide strand. We describe the evaluation of cytidine analogs for this position that stabilize an activated conformation of the enzyme-RNA complex and increase catalytic rate for deamination by the wild-type enzyme. A new crystal structure of ADAR2 bound to duplex RNA bearing a cytidine analog revealed a close contact between E488, stabilized by an additional hydrogen bond and altered charge distribution when compared to cytidine. In human cells and mouse primary liver fibroblasts, this single nucleotide modification increased directed editing yields when compared to an otherwise identical guide oligonucleotide. Our results show that modification of the guide RNA can mimic the effect of hyperactive mutants and advance the approach of recruiting endogenous ADARs for site-directed RNA editing.
- Department of Chemistry, University of California, Davis, California 95616, United States.
Organizational Affiliation: